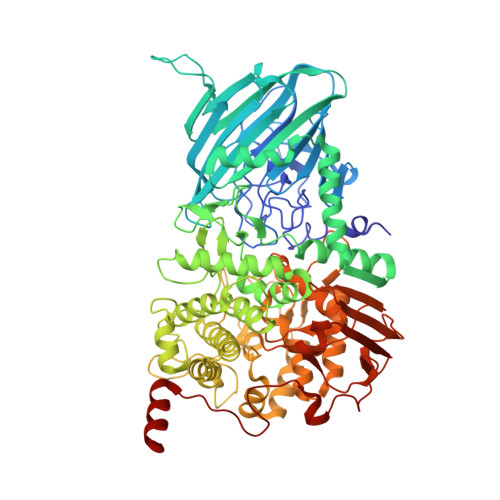
Bacteroides thetaiotaomicron generates diverse alpha-mannosidase activities through subtle evolution of a distal substrate-binding motif.
Thompson, A.J., Spears, R.J., Zhu, Y., Suits, M.D.L., Williams, S.J., Gilbert, H.J., Davies, G.J.(2018) Acta Crystallogr D Struct Biol 74: 394-404
- PubMed: 29717710
- DOI: https://doi.org/10.1107/S2059798318002942
- Primary Citation of Related Structures:
6F8Z, 6F90, 6F91, 6F92 - PubMed Abstract:
A dominant human gut microbe, the well studied symbiont Bacteroides thetaiotaomicron (Bt), is a glyco-specialist that harbors a large repertoire of genes devoted to carbohydrate processing. Despite strong similarities among them, many of the encoded enzymes have evolved distinct substrate specificities, and through the clustering of cognate genes within operons termed polysaccharide-utilization loci (PULs) enable the fulfilment of complex biological roles. Structural analyses of two glycoside hydrolase family 92 α-mannosidases, BT3130 and BT3965, together with mechanistically relevant complexes at 1.8-2.5 Å resolution reveal conservation of the global enzyme fold and core catalytic apparatus despite different linkage specificities. Structure comparison shows that Bt differentiates the activity of these enzymes through evolution of a highly variable substrate-binding region immediately adjacent to the active site. These observations unveil a genetic/biochemical mechanism through which polysaccharide-processing bacteria can evolve new and specific biochemical activities from otherwise highly similar gene products.
Organizational Affiliation:
Department of Chemistry, University of York, Heslington, York YO10 5DD, England.