Multi-target-directed ligands for treating Alzheimer's disease: Butyrylcholinesterase inhibitors displaying antioxidant and neuroprotective activities.
Knez, D., Coquelle, N., Pislar, A., Zakelj, S., Jukic, M., Sova, M., Mravljak, J., Nachon, F., Brazzolotto, X., Kos, J., Colletier, J.P., Gobec, S.(2018) Eur J Med Chem 156: 598-617
- PubMed: 30031971
- DOI: https://doi.org/10.1016/j.ejmech.2018.07.033
- Primary Citation of Related Structures:
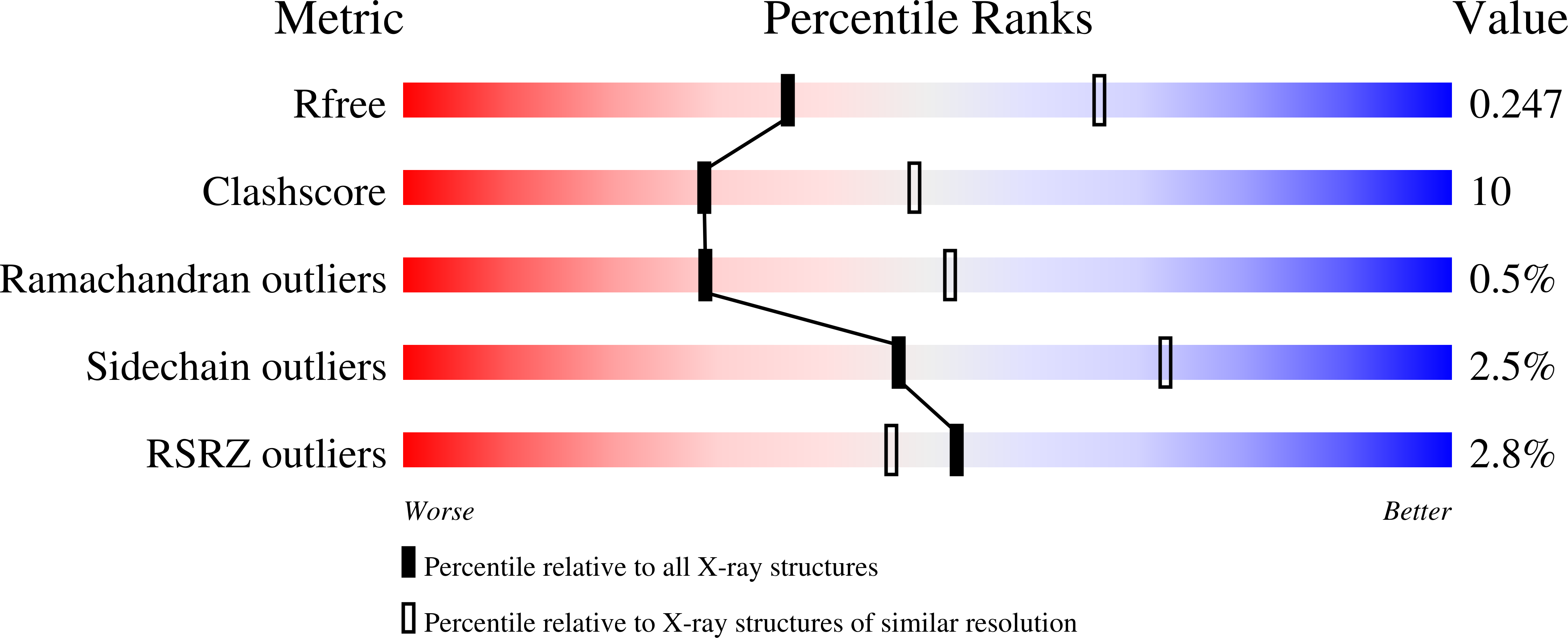
6F7Q - PubMed Abstract:
The limited clinical efficacy of current symptomatic treatment and minute effect on progression of Alzheimer's disease has shifted the research focus from single targets towards multi-target-directed ligands. Here, a potent selective inhibitor of human butyrylcholinesterase was used as the starting point to develop a new series of multifunctional ligands. A focused library of derivatives was designed and synthesised that showed both butyrylcholinesterase inhibition and good antioxidant activity as determined by the DPPH assay. The crystal structure of compound 11 in complex with butyrylcholinesterase revealed the molecular basis for its low nanomolar inhibition of butyrylcholinesterase (Ki = 1.09 ± 0.12 nM). In addition, compounds 8 and 11 show metal-chelating properties, and reduce the redox activity of chelated Cu 2+ ions in a Cu-ascorbate redox system. Compounds 8 and 11 decrease intracellular levels of reactive oxygen species, and are not substrates of the active efflux transport system, as determined in Caco2 cells. Compound 11 also protects neuroblastoma SH-SY5Y cells from toxic Aβ 1-42 species. These data indicate that compounds 8 and 11 are promising multifunctional lead ligands for treatment of Alzheimer's disease.
Organizational Affiliation:
Faculty of Pharmacy, University of Ljubljana, Aškerčeva 7, 1000, Ljubljana, Slovenia.