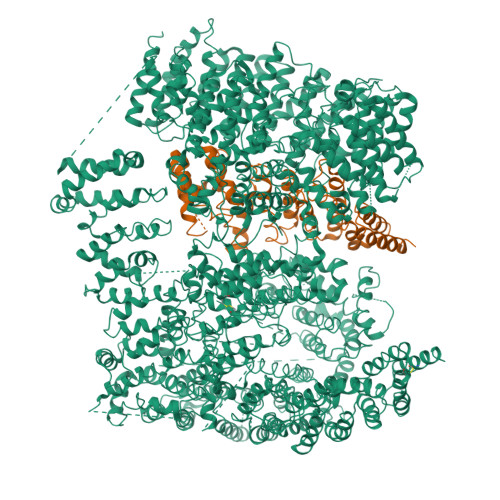
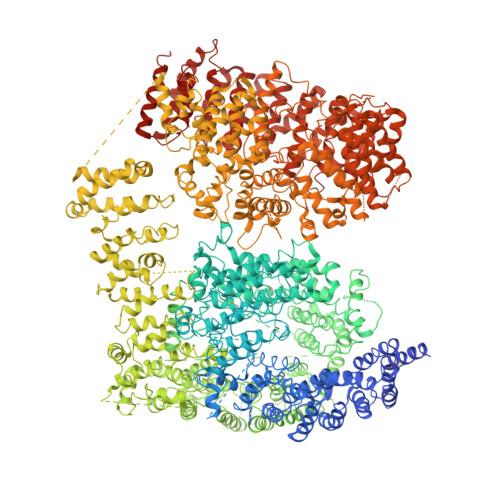
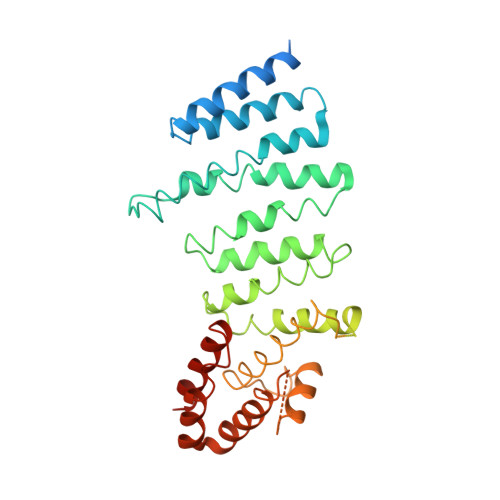
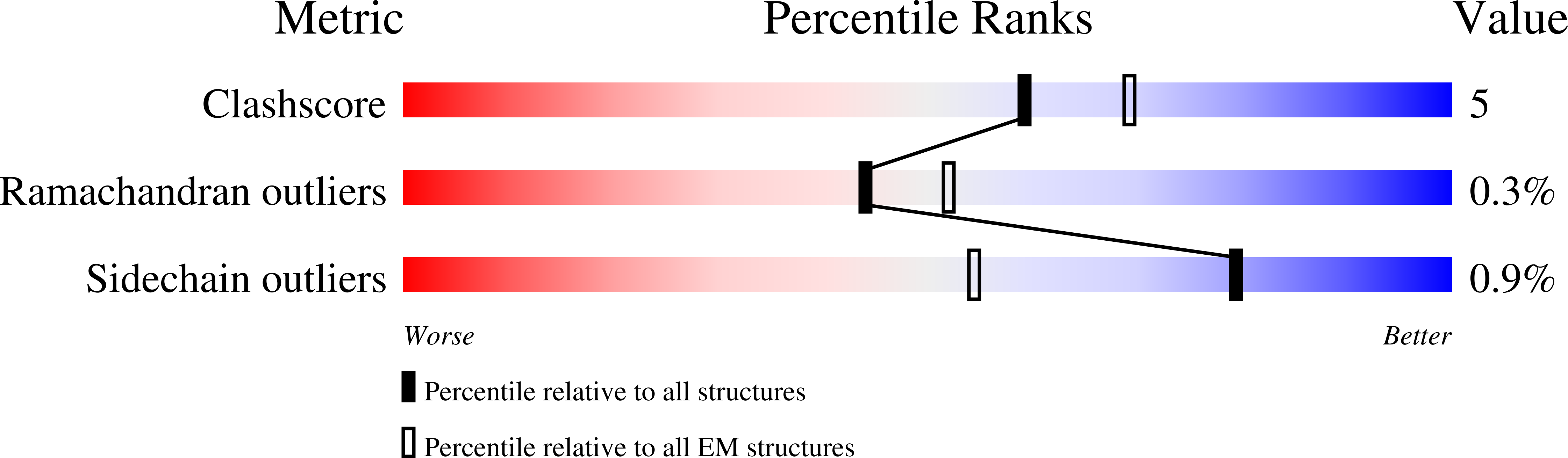The cryo-electron microscopy structure of huntingtin.
Guo, Q., Cheng, J., Seefelder, M., Engler, T., Pfeifer, G., Oeckl, P., Otto, M., Moser, F., Maurer, M., Pautsch, A., Baumeister, W., Fernandez-Busnadiego, R., Kochanek, S.(2018) Nature 555: 117-120
- PubMed: 29466333
- DOI: https://doi.org/10.1038/nature25502
- Primary Citation of Related Structures:
6EZ8 - PubMed Abstract:
Huntingtin (HTT) is a large (348 kDa) protein that is essential for embryonic development and is involved in diverse cellular activities such as vesicular transport, endocytosis, autophagy and the regulation of transcription. Although an integrative understanding of the biological functions of HTT is lacking, the large number of identified HTT interactors suggests that it serves as a protein-protein interaction hub. Furthermore, Huntington's disease is caused by a mutation in the HTT gene, resulting in a pathogenic expansion of a polyglutamine repeat at the amino terminus of HTT. However, only limited structural information regarding HTT is currently available. Here we use cryo-electron microscopy to determine the structure of full-length human HTT in a complex with HTT-associated protein 40 (HAP40; encoded by three F8A genes in humans) to an overall resolution of 4 Å. HTT is largely α-helical and consists of three major domains. The amino- and carboxy-terminal domains contain multiple HEAT (huntingtin, elongation factor 3, protein phosphatase 2A and lipid kinase TOR) repeats arranged in a solenoid fashion. These domains are connected by a smaller bridge domain containing different types of tandem repeats. HAP40 is also largely α-helical and has a tetratricopeptide repeat-like organization. HAP40 binds in a cleft and contacts the three HTT domains by hydrophobic and electrostatic interactions, thereby stabilizing the conformation of HTT. These data rationalize previous biochemical results and pave the way for improved understanding of the diverse cellular functions of HTT.
Organizational Affiliation:
Department of Molecular Structural Biology, Max Planck Institute of Biochemistry, 82152 Martinsried, Germany.