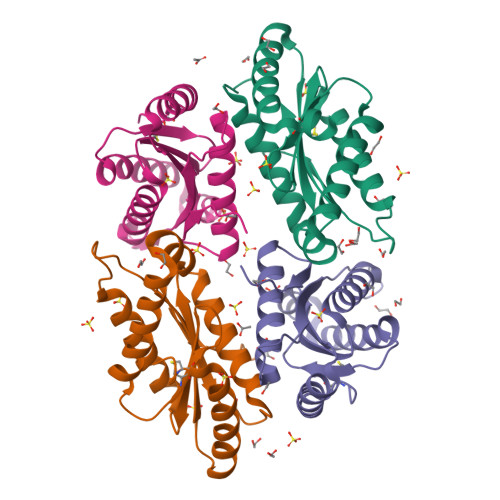
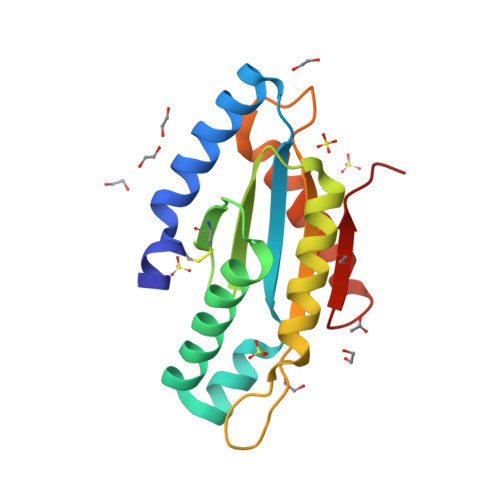
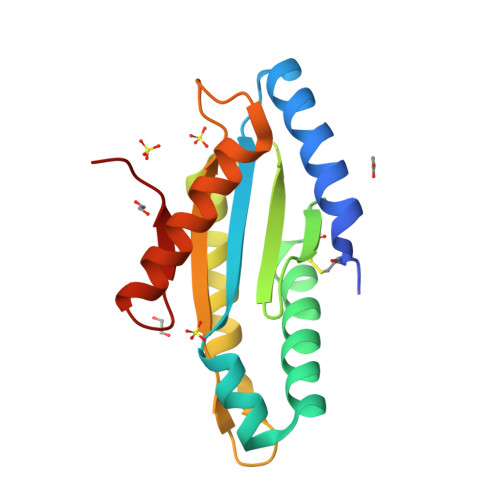
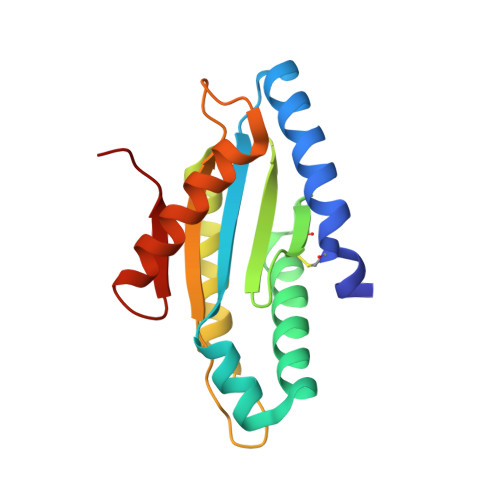
Structure of the active GGEEF domain of a diguanylate cyclase from Vibrio cholerae.
Chouhan, O.P., Roske, Y., Heinemann, U., Biswas, S.(2020) Biochem Biophys Res Commun 523: 287-292
- PubMed: 31862141
- DOI: https://doi.org/10.1016/j.bbrc.2019.11.179
- Primary Citation of Related Structures:
6EIB - PubMed Abstract:
Cyclic-di-GMP (c-di-GMP) synthesized by diguanylate cyclases has been an important and ubiquitous secondary messenger in almost all bacterial systems. In Vibrio cholerae, c-di-GMP plays an intricate role in the production of the exopolysaccharide matrix, and thereby, in biofilm formation. The formation of the surface biofilm enables the bacteria to survive in aquatic bodies, when not infecting a human host. Diguanylate cyclases are the class of enzymes which synthesize c-di-GMP from two molecules of GTP and are endowed with a GGDEF or, a GGEEF signature domain. The VC0395_0300 protein from V. cholerae, has been established as a diguanylate cyclase with a necessary role in biofilm formation. Here we present the structure of an N-terminally truncated form of VC0395_0300, which retains the active GGEEF domain for diguanylate cyclase activity but lacks 160 residues from the poorly organized N-terminal domain. X-ray diffraction data was collected from a crystal of VC0395_0300 (161-321) to a resolution of 1.9 Å. The structure displays remarkable topological similarity with diguanylate cyclases from other bacterial systems, but lacks the binding site for c-di-GMP present in its homologues. Finally, we demonstrate the ability of the truncated diguanylate cyclase VC0395_0300 (161-321) to produce c-di-GMP, and its role in biofilm formation for the bacteria.
Organizational Affiliation:
ViStA Lab, BITS, Pilani - K K Birla Goa Campus, Zuarinagar, Goa, India.