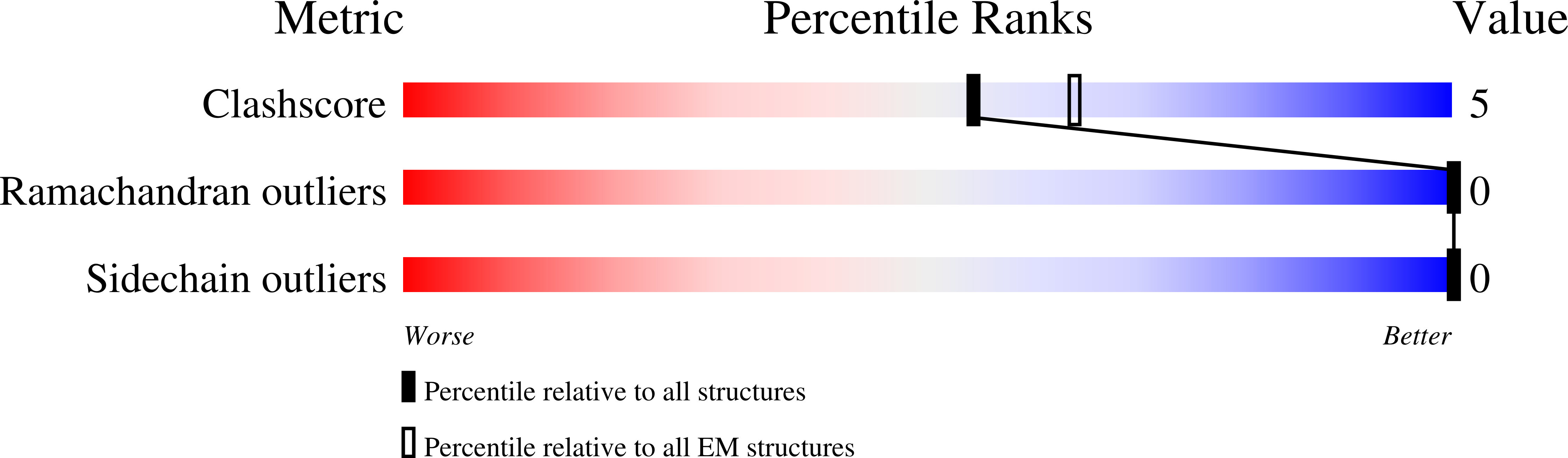Single-particle cryo-EM structure of a voltage-activated potassium channel in lipid nanodiscs.
Matthies, D., Bae, C., Toombes, G.E., Fox, T., Bartesaghi, A., Subramaniam, S., Swartz, K.J.(2018) Elife 7
- PubMed: 30109985
- DOI: https://doi.org/10.7554/eLife.37558
- Primary Citation of Related Structures:
6EBK, 6EBL, 6EBM - PubMed Abstract:
Voltage-activated potassium (Kv) channels open to conduct K + ions in response to membrane depolarization, and subsequently enter non-conducting states through distinct mechanisms of inactivation. X-ray structures of detergent-solubilized Kv channels appear to have captured an open state even though a non-conducting C-type inactivated state would predominate in membranes in the absence of a transmembrane voltage. However, structures for a voltage-activated ion channel in a lipid bilayer environment have not yet been reported. Here we report the structure of the Kv1.2-2.1 paddle chimera channel reconstituted into lipid nanodiscs using single-particle cryo-electron microscopy. At a resolution of ~3 Å for the cytosolic domain and ~4 Å for the transmembrane domain, the structure determined in nanodiscs is similar to the previously determined X-ray structure. Our findings show that large differences in structure between detergent and lipid bilayer environments are unlikely, and enable us to propose possible structural mechanisms for C-type inactivation.
Organizational Affiliation:
Laboratory of Cell Biology, Center for Cancer Research, National Cancer Institute, National Institutes of Health, Bethesda, United States.