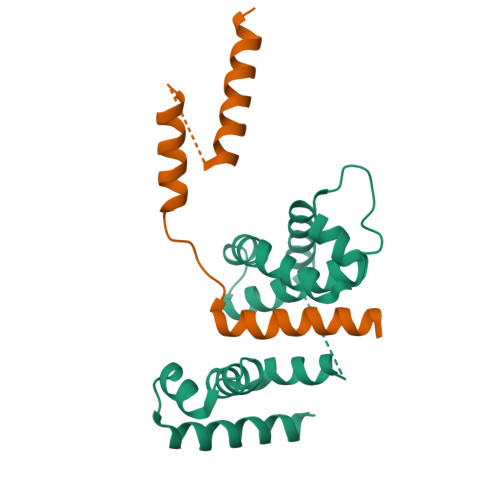
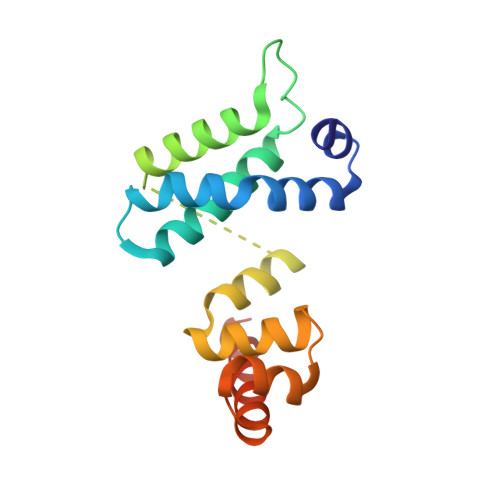
The crystal structure of the RsbN-sigma BldN complex from Streptomyces venezuelae defines a new structural class of anti-sigma factor.
Schumacher, M.A., Bush, M.J., Bibb, M.J., Ramos-Leon, F., Chandra, G., Zeng, W., Buttner, M.J.(2018) Nucleic Acids Res 46: 7405-7417
- PubMed: 29905823
- DOI: https://doi.org/10.1093/nar/gky493
- Primary Citation of Related Structures:
6DXO - PubMed Abstract:
Streptomyces are filamentous bacteria with a complex developmental life cycle characterized by the formation of spore-forming aerial hyphae. Transcription of the chaplin and rodlin genes, which are essential for aerial hyphae production, is directed by the extracytoplasmic function (ECF) σ factor BldN, which is in turn controlled by an anti-σ factor, RsbN. RsbN shows no sequence similarity to known anti-σ factors and binds and inhibits BldN in an unknown manner. Here we describe the 2.23 Å structure of the RsbN-BldN complex. The structure shows that BldN harbors σ2 and σ4 domains that are individually similar to other ECF σ domains, which bind -10 and -35 promoter regions, respectively. The anti-σ RsbN consists of three helices, with α3 forming a long helix embraced between BldN σ2 and σ4 while RsbN α1-α2 dock against σ4 in a manner that would block -35 DNA binding. RsbN binding also freezes BldN in a conformation inactive for simultaneous -10 and -35 promoter interaction and RNAP binding. Strikingly, RsbN is structurally distinct from previously solved anti-σ proteins. Thus, these data characterize the molecular determinants controlling a central Streptomyces developmental switch and reveal RsbN to be the founding member of a new structural class of anti-σ factor.
Organizational Affiliation:
Department of Biochemistry, Duke University School of Medicine, Durham, NC 27710, USA.