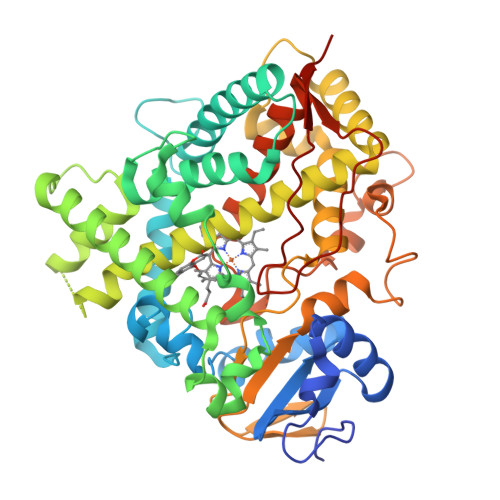
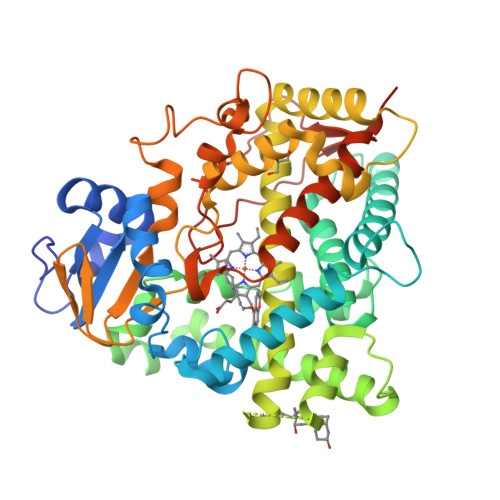
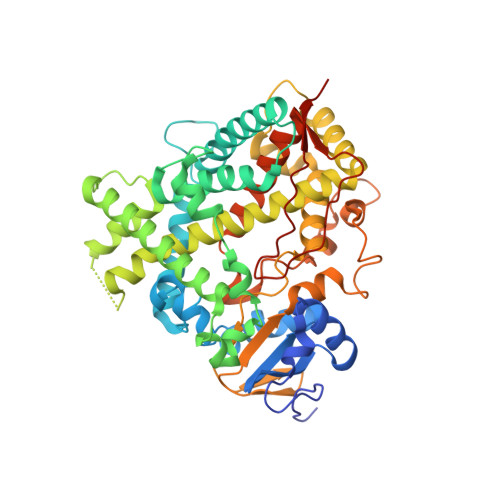
Structures of human cytochrome P450 1A1 with bergamottin and erlotinib reveal active-site modifications for binding of diverse ligands.
Bart, A.G., Scott, E.E.(2018) J Biological Chem 293: 19201-19210
- PubMed: 30254074
- DOI: https://doi.org/10.1074/jbc.RA118.005588
- Primary Citation of Related Structures:
6DWM, 6DWN - PubMed Abstract:
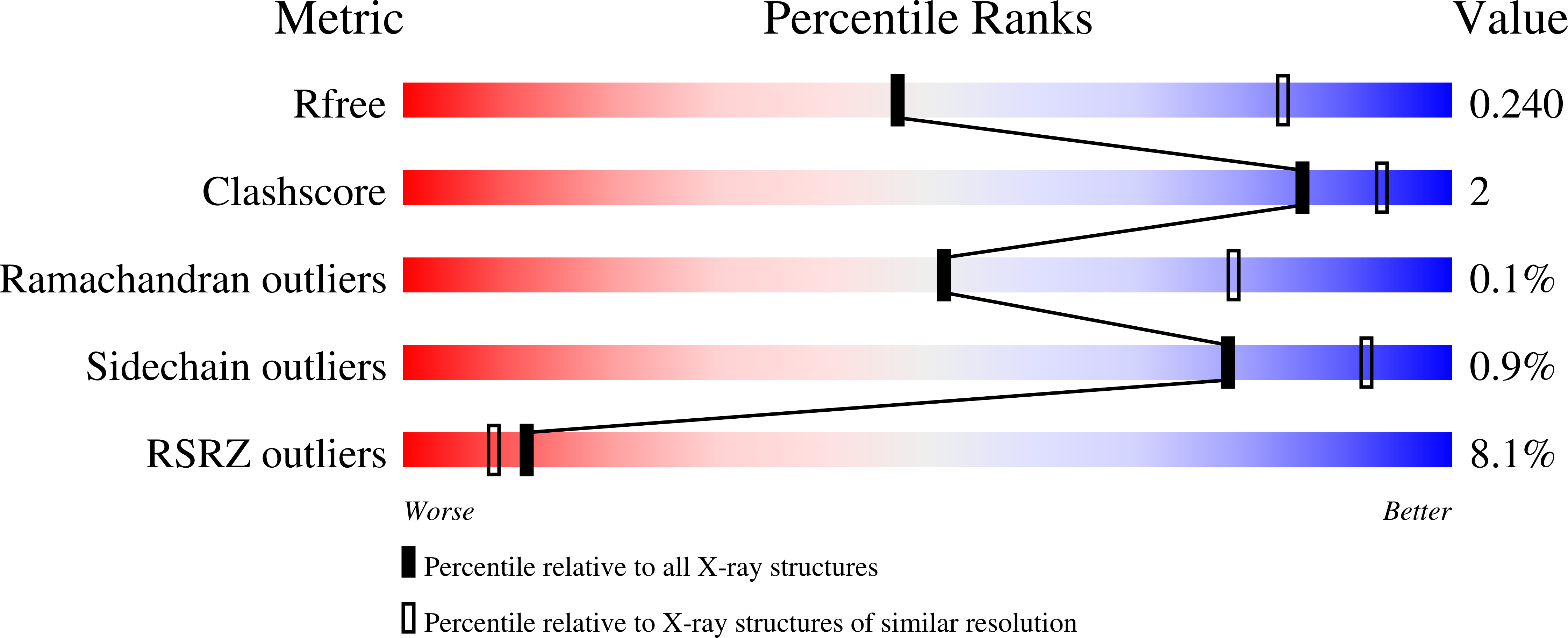
Human cytochrome P450 1A1 (CYP1A1) is an extrahepatic enzyme involved in the monooxygenation of structurally diverse compounds ranging from natural products to drugs and protoxins. Because CYP1A1 has a role in human carcinogenesis, inhibiting its activity may potentially aid in cancer chemoprevention, whereas utilizing CYP1A1's oxidative activity could help selectively activate anticancer prodrugs. Such potential therapeutic purposes require detailed knowledge of CYP1A1's interactions with potential ligands. Known CYP1A1 ligands also vary substantially in size, and it has not been apparent from a single existing CYP1A1 structure how larger, structurally diverse ligands are accommodated within the enclosed active site. Here, two new X-ray structures with the natural product furanocoumarin bergamottin (at 2.85 Å resolution) and the lung cancer drug erlotinib (3.0 Å) revealed binding orientations consistent with the formation of innocuous metabolites and of toxic metabolites, respectively. They also disclosed local changes in the roof of the active site that enlarge the active site and ultimately form a channel to the protein exterior. Although further structural modifications would be required to accommodate the largest CYP1A1 ligands, knowing which components of the active site are malleable provides powerful information for those attempting to use computational approaches to predict compound binding and substrate metabolism by this clinically relevant monooxygenase.
Organizational Affiliation:
From the Program in Biophysics and.