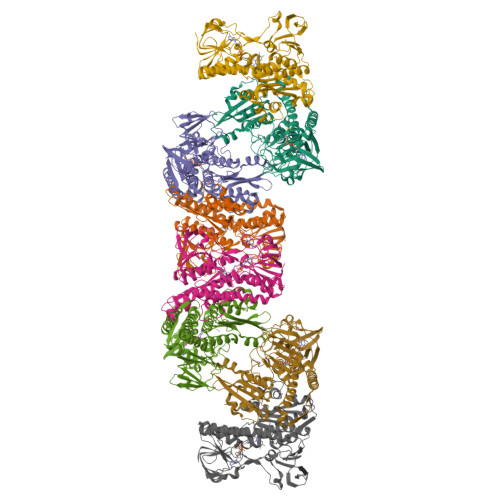
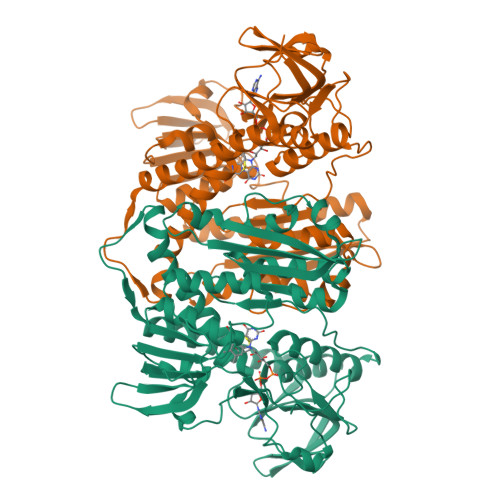
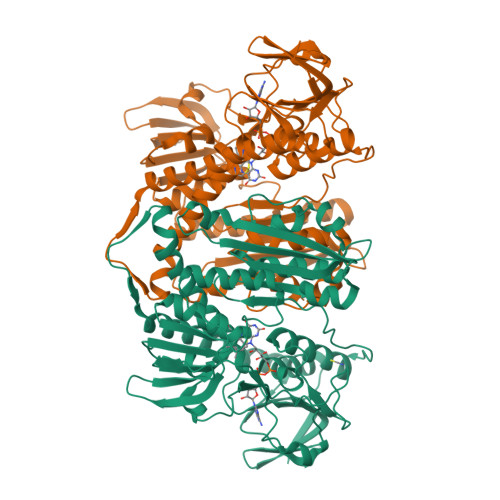
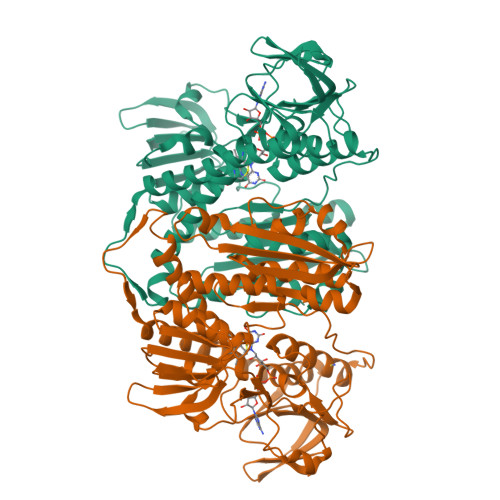
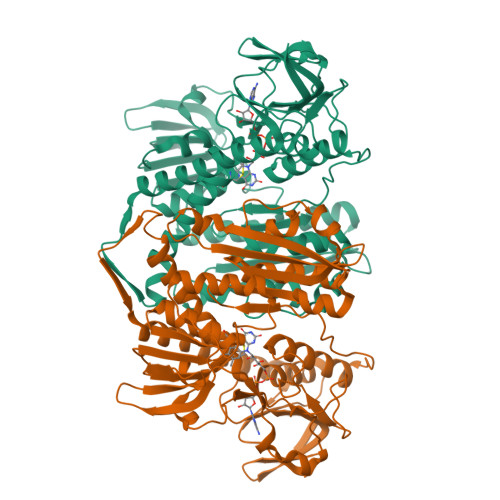
The structure and activity of the glutathione reductase from Streptococcus pneumoniae.
Sikanyika, M., Aragao, D., McDevitt, C.A., Maher, M.J.(2019) Acta Crystallogr F Struct Biol Commun 75: 54-61
- PubMed: 30605126
- DOI: https://doi.org/10.1107/S2053230X18016527
- Primary Citation of Related Structures:
6DU7 - PubMed Abstract:
The glutathione reductase (GR) from Streptococcus pneumoniae is a flavoenzyme that catalyzes the reduction of oxidized glutathione (GSSG) to its reduced form (GSH) in the cytoplasm of this bacterium. The maintenance of an intracellular pool of GSH is critical for the detoxification of reactive oxygen and nitrogen species and for intracellular metal tolerance to ions such as zinc. Here, S. pneumoniae GR (SpGR) was overexpressed and purified and its crystal structure determined at 2.56 Å resolution. SpGR shows overall structural similarity to other characterized GRs, with a dimeric structure that includes an antiparallel β-sheet at the dimer interface. This observation, in conjunction with comparisons with the interface structures of other GR enzymes, allows the classification of these enzymes into three classes. Analyses of the kinetic properties of SpGR revealed a significantly higher value for K m(GSSG) (231.2 ± 24.7 µM) in comparison to other characterized GR enzymes.
Organizational Affiliation:
Department of Biochemistry and Genetics, La Trobe Institute for Molecular Science, La Trobe University, Melbourne 3086, Australia.