De novo design of potent and selective mimics of IL-2 and IL-15.
Silva, D.A., Yu, S., Ulge, U.Y., Spangler, J.B., Jude, K.M., Labao-Almeida, C., Ali, L.R., Quijano-Rubio, A., Ruterbusch, M., Leung, I., Biary, T., Crowley, S.J., Marcos, E., Walkey, C.D., Weitzner, B.D., Pardo-Avila, F., Castellanos, J., Carter, L., Stewart, L., Riddell, S.R., Pepper, M., Bernardes, G.J.L., Dougan, M., Garcia, K.C., Baker, D.(2019) Nature 565: 186-191
- PubMed: 30626941
- DOI: https://doi.org/10.1038/s41586-018-0830-7
- Primary Citation of Related Structures:
6DG5, 6DG6 - PubMed Abstract:
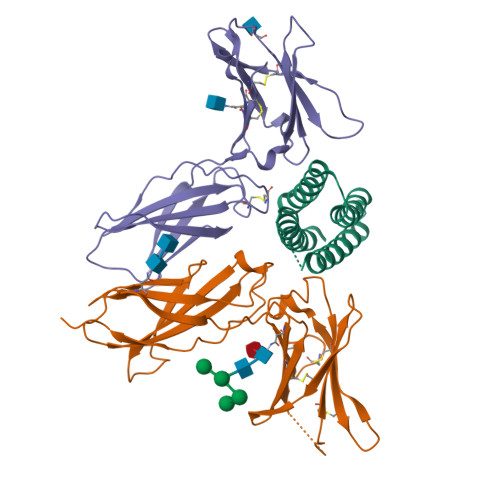
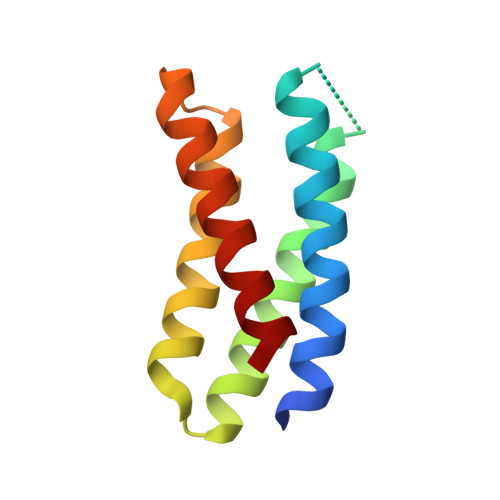
We describe a de novo computational approach for designing proteins that recapitulate the binding sites of natural cytokines, but are otherwise unrelated in topology or amino acid sequence. We use this strategy to design mimics of the central immune cytokine interleukin-2 (IL-2) that bind to the IL-2 receptor βγ c heterodimer (IL-2Rβγ c ) but have no binding site for IL-2Rα (also called CD25) or IL-15Rα (also known as CD215). The designs are hyper-stable, bind human and mouse IL-2Rβγ c with higher affinity than the natural cytokines, and elicit downstream cell signalling independently of IL-2Rα and IL-15Rα. Crystal structures of the optimized design neoleukin-2/15 (Neo-2/15), both alone and in complex with IL-2Rβγ c , are very similar to the designed model. Neo-2/15 has superior therapeutic activity to IL-2 in mouse models of melanoma and colon cancer, with reduced toxicity and undetectable immunogenicity. Our strategy for building hyper-stable de novo mimetics could be applied generally to signalling proteins, enabling the creation of superior therapeutic candidates.
Organizational Affiliation:
Institute for Protein Design, University of Washington, Seattle, WA, USA. dadriano@uw.edu.