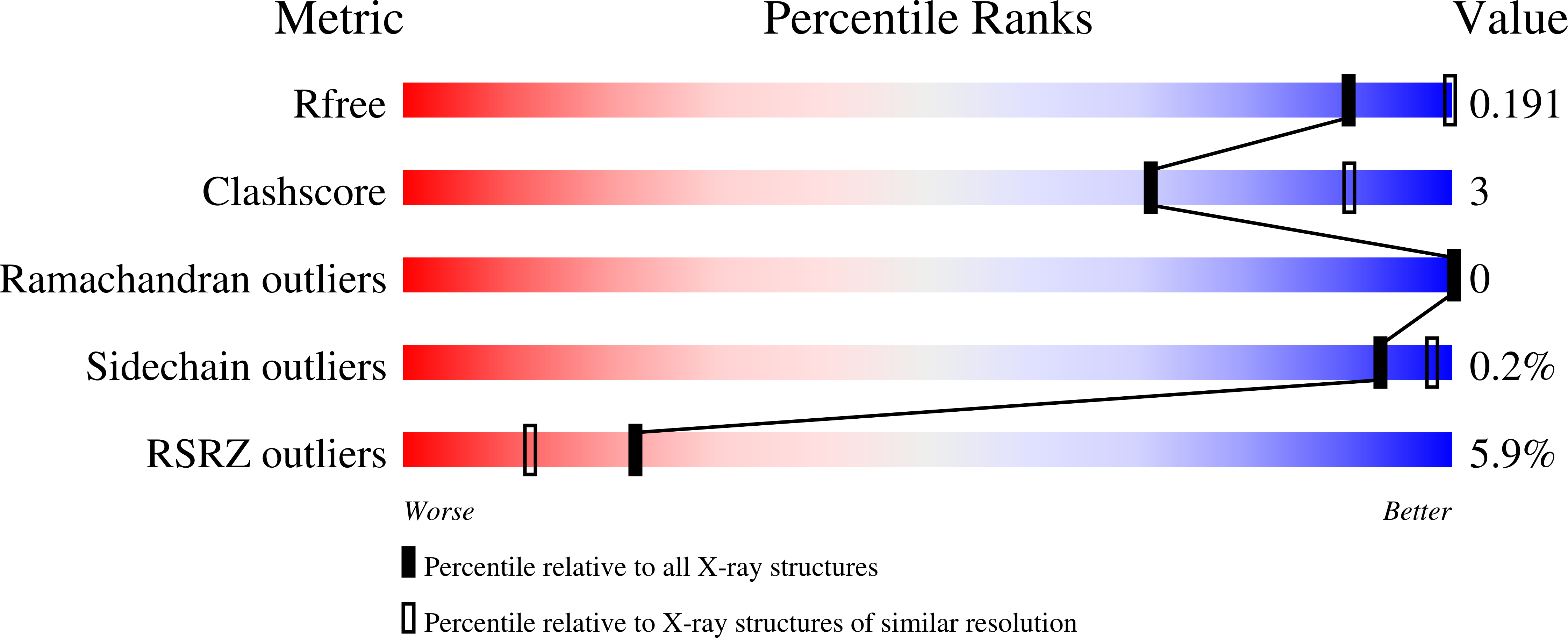
Commercial AHAS-inhibiting herbicides are promising drug leads for the treatment of human fungal pathogenic infections.
Garcia, M.D., Chua, S.M.H., Low, Y.S., Lee, Y.T., Agnew-Francis, K., Wang, J.G., Nouwens, A., Lonhienne, T., Williams, C.M., Fraser, J.A., Guddat, L.W.(2018) Proc Natl Acad Sci U S A 115: E9649-E9658
- PubMed: 30249642
- DOI: https://doi.org/10.1073/pnas.1809422115
- Primary Citation of Related Structures:
6DEK, 6DEL, 6DEM, 6DEN, 6DEO, 6DEP, 6DEQ, 6DER, 6DES - PubMed Abstract:
The increased prevalence of drug-resistant human pathogenic fungal diseases poses a major threat to global human health. Thus, new drugs are urgently required to combat these infections. Here, we demonstrate that acetohydroxyacid synthase (AHAS), the first enzyme in the branched-chain amino acid biosynthesis pathway, is a promising new target for antifungal drug discovery. First, we show that several AHAS inhibitors developed as commercial herbicides are powerful accumulative inhibitors of Candida albicans AHAS ( K i values as low as 800 pM) and have determined high-resolution crystal structures of this enzyme in complex with several of these herbicides. In addition, we have demonstrated that chlorimuron ethyl (CE), a member of the sulfonylurea herbicide family, has potent antifungal activity against five different Candida species and Cryptococcus neoformans (with minimum inhibitory concentration, 50% values as low as 7 nM). Furthermore, in these assays, we have shown CE and itraconazole (a P450 inhibitor) can act synergistically to further improve potency. Finally, we show in Candida albicans -infected mice that CE is highly effective in clearing pathogenic fungal burden in the lungs, liver, and spleen, thus reducing overall mortality rates. Therefore, in view of their low toxicity to human cells, AHAS inhibitors represent a new class of antifungal drug candidates.
Organizational Affiliation:
School of Chemistry and Molecular Biosciences, The University of Queensland, Brisbane, QLD 4072, Australia.