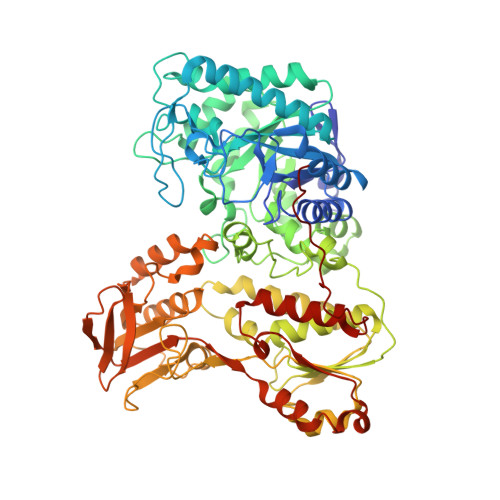
Structure of Rhizobium sp. 4-9 histamine dehydrogenase and analysis of the electron transfer pathway to an abiological electron acceptor.
Goyal, P., Deay 3rd, D., Seibold, S., Candido, A.C.L., Lovell, S., Battaile, K.P., Wilson, G.S., Richter, M.L., Petillo, P.A.(2023) Arch Biochem Biophys : 109612-109612
- PubMed: 37146865
- DOI: https://doi.org/10.1016/j.abb.2023.109612
- Primary Citation of Related Structures:
6DE6 - PubMed Abstract:
Histamine dehydrogenase from the gram-negative bacterium Rhizobium sp. 4-9 (HaDHR) is a member of a small family of dehydrogenases containing a covalently attached FMN, and the only member so far identified to date that does not exhibit substrate inhibition. In this study, we present the 2.1 Å resolution crystal structure of HaDHR. This new structure allowed for the identification of the internal electron transfer pathway to abiological ferrocene-based mediators. Alanine 437 was identified as the exit point of electrons from the Fe 4 S 4 cluster. The enzyme was modified with a Ser436Cys mutation to facilitate covalent attachment of a ferrocene moiety. When modified with Fc-maleimide, this new construct demonstrated direct electron transfer from the enzyme to a gold electrode in a histamine concentration-dependent manner without the need for any additional electron mediators.
Organizational Affiliation:
Department of Molecular Biosciences, University of Kansas, Lawrence, KS, 66045, USA.