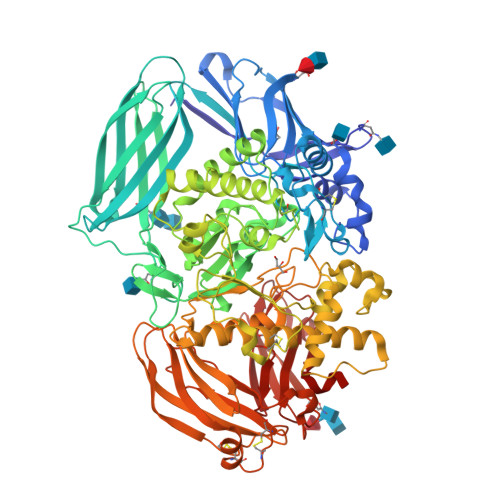
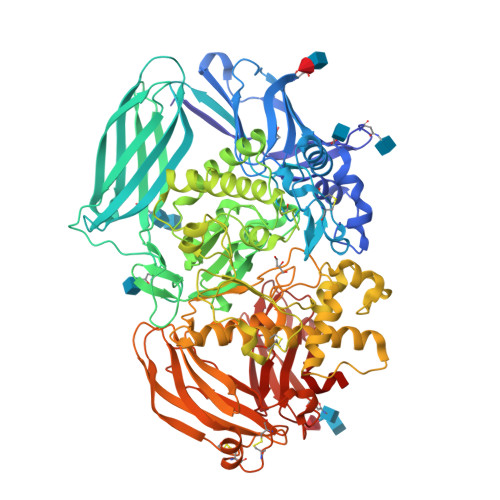
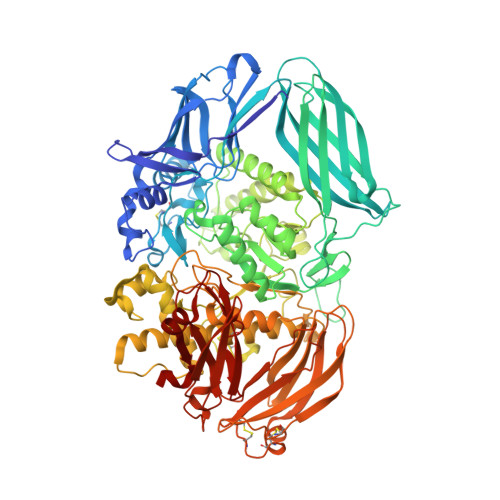
The structure of mammalian beta-mannosidase provides insight into beta-mannosidosis and nystagmus.
Gytz, H., Liang, J., Liang, Y., Gorelik, A., Illes, K., Nagar, B.(2019) FEBS J 286: 1319-1331
- PubMed: 30552791
- DOI: https://doi.org/10.1111/febs.14731
- Primary Citation of Related Structures:
6DDT, 6DDU - PubMed Abstract:
β-Mannosidase is a lysosomal enzyme from the glycosyl hydrolase family 2 that cleaves the single β(1-4)-linked mannose at the nonreducing end of N-glycosylated proteins, and plays an important role in the polysaccharide degradation pathway. Mutations in the MANBA gene, which encodes the β-mannosidase, can lead to the lysosomal storage disease β-mannosidosis, as well as nystagmus, an eye condition characterized by involuntary eye movements. Here, we present the first structures of a mammalian β-mannosidase in both the apo- and mannose-bound forms. The structure is similar to previously determined β-mannosidase structures with regard to domain organization and fold, however, there are important differences that underlie substrate specificity between species. Additionally, in contrast to most other ligand-bound β-mannosidases from bacterial and fungal sources where bound sugars were in a boat-like conformation, we find the mannose in the chair conformation. Evaluation of known disease mutations in the MANBA gene provides insight into their impact on disease phenotypes. Together, these results will be important for the design of therapeutics for treating diseases caused by β-mannosidase deficiency. DATABASE: Structural data are available in the Protein Data Bank under the accession numbers 6DDT and 6DDU.
Organizational Affiliation:
Department of Biochemistry, Groupe de Recherche Axé sur la Structure des Protéines, McGill University, Montreal, Canada.