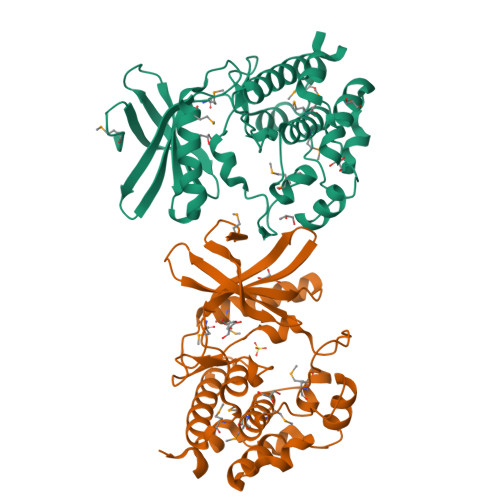
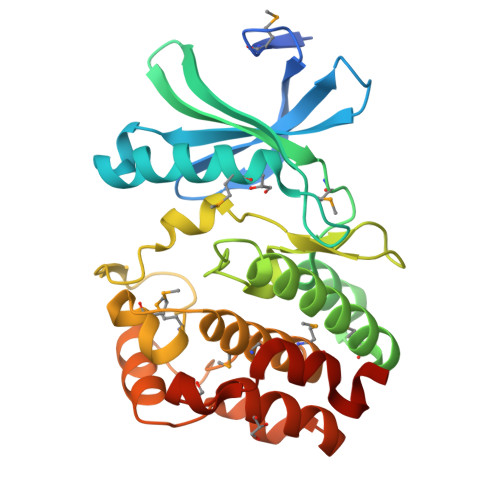
Crystal structure of the kinase domain of WNK1, a kinase that causes a hereditary form of hypertension.
Min, X., Lee, B.H., Cobb, M.H., Goldsmith, E.J.(2004) Structure 12: 1303-1311
- PubMed: 15242606
- DOI: https://doi.org/10.1016/j.str.2004.04.014
- Primary Citation of Related Structures:
6CN9 - PubMed Abstract:
WNK kinases comprise a small group of unique serine/threonine protein kinases that have been genetically linked to pseudohypoaldosteronism type II, an autosomal dominant form of hypertension. Here we present the structure of the kinase domain of WNK1 at 1.8 A resolution, solved in a low activity conformation. A lysine residue (Lys-233) is found in the active site emanating from strand beta2 rather than strand beta3 as in other protein kinases. The activation loop adopts a unique well-folded inactive conformation. The conformations of the P+1 specificity pocket, the placement of the conserved active site threonine (Thr-386), and the exterior placement of helix C, contribute to the low activity state. By homology modeling, we identified two hydrophobic residues in the substrate-binding groove that contribute to substrate specificity. The structure of the WNK1 catalytic domain, with its unique active site, may help in the design of therapeutic reagents for the treatment of hypertension.
Organizational Affiliation:
Department of Biochemistry, University of Texas Southwestern Medical Center at Dallas, 5323 Harry Hines Boulevard, Dallas, TX 75390, USA.