Structural Basis for Teneurin Function in Circuit-Wiring: A Toxin Motif at the Synapse.
Li, J., Shalev-Benami, M., Sando, R., Jiang, X., Kibrom, A., Wang, J., Leon, K., Katanski, C., Nazarko, O., Lu, Y.C., Sudhof, T.C., Skiniotis, G., Arac, D.(2018) Cell 173: 735-748.e15
- PubMed: 29677516
- DOI: https://doi.org/10.1016/j.cell.2018.03.036
- Primary Citation of Related Structures:
6CMX - PubMed Abstract:
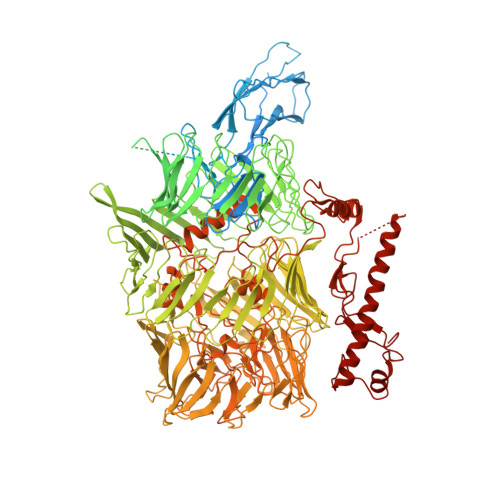
Teneurins (TENs) are cell-surface adhesion proteins with critical roles in tissue development and axon guidance. Here, we report the 3.1-Å cryoelectron microscopy structure of the human TEN2 extracellular region (ECR), revealing a striking similarity to bacterial Tc-toxins. The ECR includes a large β barrel that partially encapsulates a C-terminal domain, which emerges to the solvent through an opening in the mid-barrel region. An immunoglobulin (Ig)-like domain seals the bottom of the barrel while a β propeller is attached in a perpendicular orientation. We further show that an alternatively spliced region within the β propeller acts as a switch to regulate trans-cellular adhesion of TEN2 to latrophilin (LPHN), a transmembrane receptor known to mediate critical functions in the central nervous system. One splice variant activates trans-cellular signaling in a LPHN-dependent manner, whereas the other induces inhibitory postsynaptic differentiation. These results highlight the unusual structural organization of TENs giving rise to their multifarious functions.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, The University of Chicago, Chicago, IL 60637, USA; Grossman Institute for Neuroscience, Quantitative Biology and Human Behavior, The University of Chicago, Chicago, IL 60637, USA.