The binding of DCC-P3 motif and FAK-FAT domain mediates the initial step of netrin-1/DCC signaling for axon attraction.
Xu, S., Liu, Y., Li, X., Liu, Y., Meijers, R., Zhang, Y., Wang, J.H.(2018) Cell Discov 4: 8-8
- PubMed: 29479476
- DOI: https://doi.org/10.1038/s41421-017-0008-8
- Primary Citation of Related Structures:
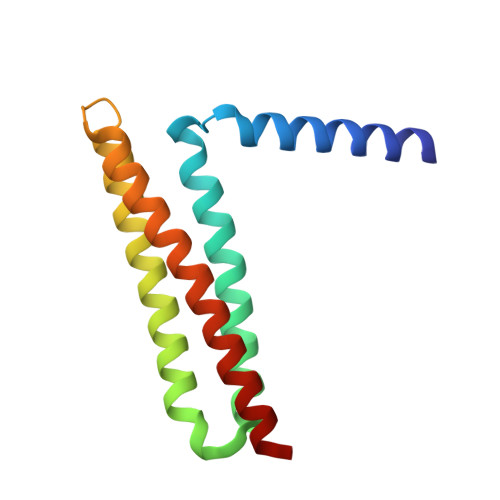
6BZ3 - PubMed Abstract:
Netrin-1 plays a key role in axon guidance through binding to its receptor, Deleted in Colorectal Cancer (DCC). The initial step of signaling inside the cell after netrin-1/DCC ligation is the binding of DCC cytoplasmic P3 motif to focal adhesion targeting (FAT) domain of focal adhesion kinase (FAK). Here we report the crystal structure of P3/FAT complex. The helical P3 peptide interacts with a helix-swapped FAT dimer in a 2:2 ratio. Dimeric FAT binding is P3-specific and stabilized by a calcium ion. Biochemical studies showed that DCC-P3 motif and calcium ion could facilitate FAT dimerization in solution. Axon guidance assays confirm that the DCC/FAK complex is essential for netrin-1-induced chemoattraction. We propose that netrin-1/DCC engagement creates a small cluster of P3/FAT for FAK recruitment close to the cell membrane, which exerts a concerted effect with PIP2 for FAK signaling. We also compare P3/FAT binding with paxillin/FAT binding and discuss their distinct recognition specificity on a common FAT domain for axon attraction versus integrin signaling, respectively.
Organizational Affiliation:
1College of Life Science and Technology, Huazhong Agricultural University, Wuhan, 430070 China.