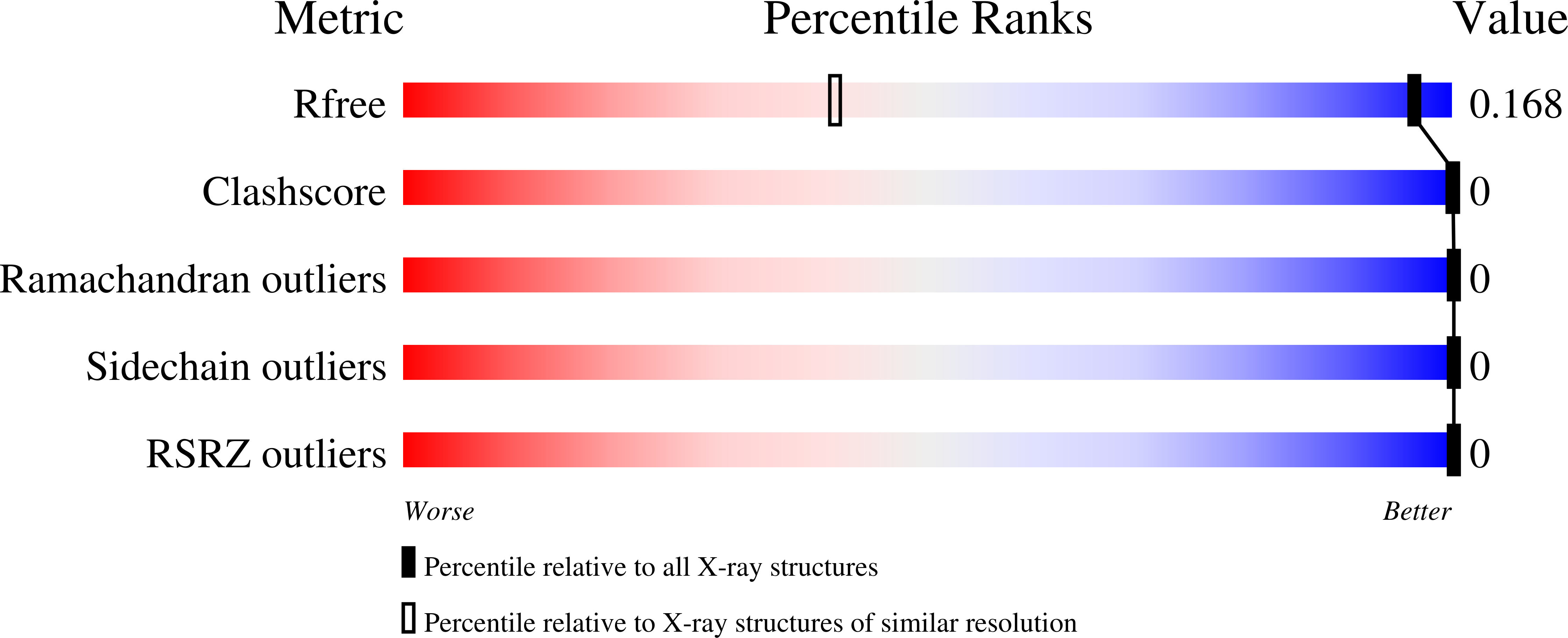
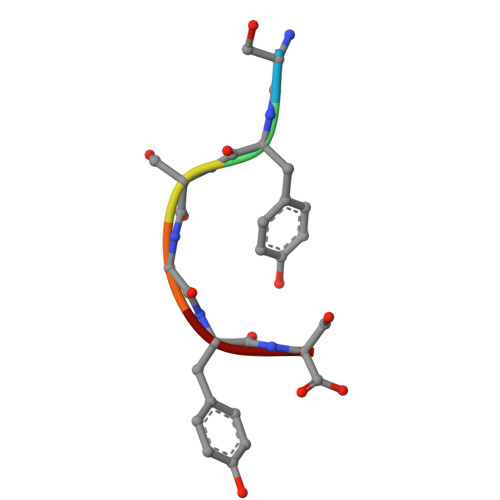
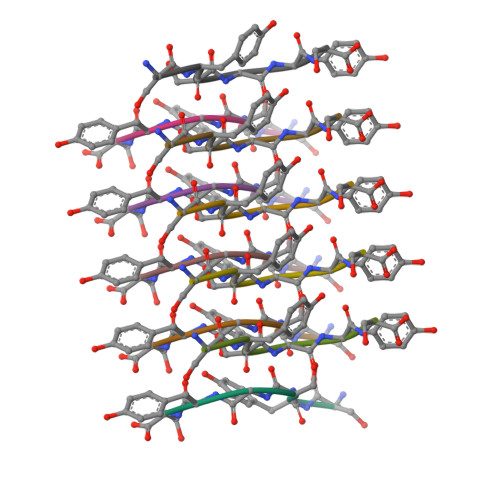
Atomic structures of low-complexity protein segments reveal kinked beta sheets that assemble networks.
Hughes, M.P., Sawaya, M.R., Boyer, D.R., Goldschmidt, L., Rodriguez, J.A., Cascio, D., Chong, L., Gonen, T., Eisenberg, D.S.(2018) Science 359: 698-701
- PubMed: 29439243
- DOI: https://doi.org/10.1126/science.aan6398
- Primary Citation of Related Structures:
6BWZ, 6BXV, 6BXX, 6BZM, 6BZP - PubMed Abstract:
Subcellular membraneless assemblies are a reinvigorated area of study in biology, with spirited scientific discussions on the forces between the low-complexity protein domains within these assemblies. To illuminate these forces, we determined the atomic structures of five segments from protein low-complexity domains associated with membraneless assemblies. Their common structural feature is the stacking of segments into kinked β sheets that pair into protofilaments. Unlike steric zippers of amyloid fibrils, the kinked sheets interact weakly through polar atoms and aromatic side chains. By computationally threading the human proteome on our kinked structures, we identified hundreds of low-complexity segments potentially capable of forming such interactions. These segments are found in proteins as diverse as RNA binders, nuclear pore proteins, and keratins, which are known to form networks and localize to membraneless assemblies.
Organizational Affiliation:
Department of Biological Chemistry and Department of Chemistry and Biochemistry, University of California Los Angeles (UCLA), Howard Hughes Medical Institute (HHMI), UCLA-Department of Energy (DOE) Institute for Genomics and Proteomics, Los Angeles, CA 90095, USA.