Discovery of Trifluoromethyl Glycol Carbamates as Potent and Selective Covalent Monoacylglycerol Lipase (MAGL) Inhibitors for Treatment of Neuroinflammation.
McAllister, L.A., Butler, C.R., Mente, S., O'Neil, S.V., Fonseca, K.R., Piro, J.R., Cianfrogna, J.A., Foley, T.L., Gilbert, A.M., Harris, A.R., Helal, C.J., Johnson, D.S., Montgomery, J.I., Nason, D.M., Noell, S., Pandit, J., Rogers, B.N., Samad, T.A., Shaffer, C.L., da Silva, R.G., Uccello, D.P., Webb, D., Brodney, M.A.(2018) J Med Chem 61: 3008-3026
- PubMed: 29498843
- DOI: https://doi.org/10.1021/acs.jmedchem.8b00070
- Primary Citation of Related Structures:
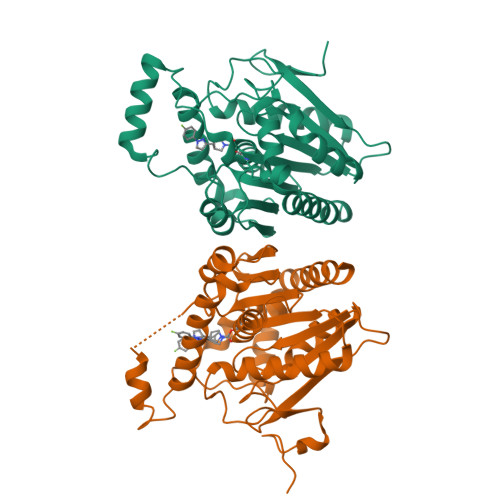
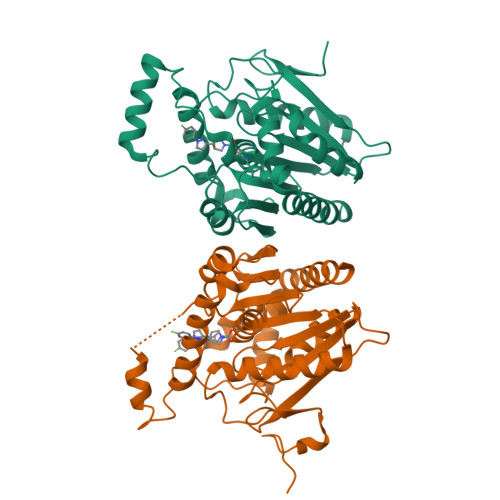
6BQ0 - PubMed Abstract:
Monoacylglycerol lipase (MAGL) inhibition provides a potential treatment approach to neuroinflammation through modulation of both the endocannabinoid pathway and arachidonoyl signaling in the central nervous system (CNS). Herein we report the discovery of compound 15 (PF-06795071), a potent and selective covalent MAGL inhibitor, featuring a novel trifluoromethyl glycol leaving group that confers significant physicochemical property improvements as compared with earlier inhibitor series with more lipophilic leaving groups. The design strategy focused on identifying an optimized leaving group that delivers MAGL potency, serine hydrolase selectivity, and CNS exposure while simultaneously reducing log D, improving solubility, and minimizing chemical lability. Compound 15 achieves excellent CNS exposure, extended 2-AG elevation effect in vivo, and decreased brain inflammatory markers in response to an inflammatory challenge.
Organizational Affiliation:
Pfizer Worldwide Research and Development , 610 Main Street , Cambridge , Massachusetts 02139 , United States.