Structural Mechanism of the Oxygenase JMJD6 Recognition by the Extraterminal (ET) Domain of BRD4.
Konuma, T., Yu, D., Zhao, C., Ju, Y., Sharma, R., Ren, C., Zhang, Q., Zhou, M.M., Zeng, L.(2017) Sci Rep 7: 16272-16272
- PubMed: 29176719
- DOI: https://doi.org/10.1038/s41598-017-16588-8
- Primary Citation of Related Structures:
6BNH - PubMed Abstract:
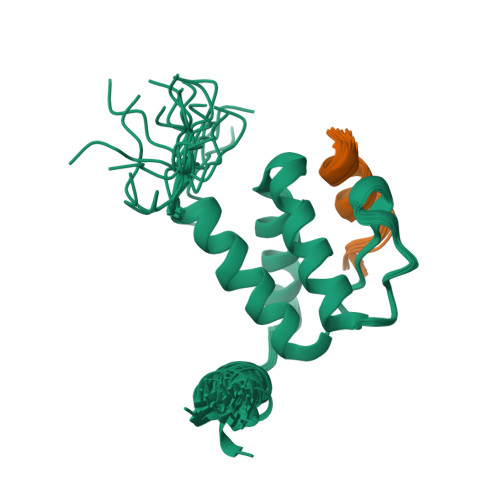
Jumonji domain-containing protein 6 (JMJD6) is a member of the Jumonji C family of Fe(II) and 2-oxoglutarate (2OG) dependent oxygenases. It possesses unique bi-functional oxygenase activities, acting as both an arginine demethylase and a lysyl-hydroxylase. JMJD6 has been reported to be over-expressed in oral, breast, lung, and colon cancers and plays important roles in regulation of transcription through interactions with transcription regulator BRD4, histones, U2AF65, Luc7L3, and SRSF11. Here, we report a structural mechanism revealed by NMR of JMJD6 recognition by the extraterminal (ET) domain of BRD4 in that a JMJD6 peptide (Lys84-Asn96) adapts an α-helix when bound to the ET domain. This intermolecular recognition is established through JMJD6 interactions with the conserved hydrophobic core of the ET domain, and reinforced by electrostatic interactions of JMJD6 with residues in the inter-helical α1-α2 loop of the ET domain. Notably, this mode of ligand recognition is different from that of ET domain recognition of NSD3, LANA of herpesvirus, and integrase of MLV, which involves formation of an intermolecular amphipathic two- or three- strand antiparallel β sheet. Furthermore, we demonstrate that the association between the BRD4 ET domain and JMJD6 likely requires a protein conformational change induced by single-stranded RNA binding.
Organizational Affiliation:
Department of Pharmacological Sciences, Icahn School of Medicine at Mount Sinai, New York, NY, 10029, USA.