Structural and functional characterisation of the entry point to pyocyanin biosynthesis inPseudomonas aeruginosadefines a new 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase subclass.
Sterritt, O.W., Lang, E.J.M., Kessans, S.A., Ryan, T.M., Demeler, B., Jameson, G.B., Parker, E.J.(2018) Biosci Rep 38
- PubMed: 30242059
- DOI: https://doi.org/10.1042/BSR20181605
- Primary Citation of Related Structures:
6BMC - PubMed Abstract:
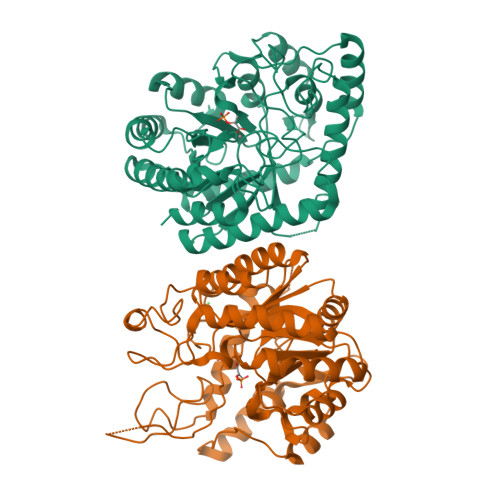
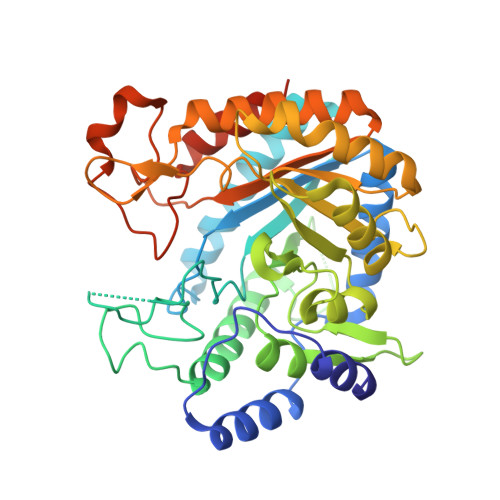
In Pseudomonas aeruginosa ( Pae ), the shikimate pathway end product, chorismate, serves as the last common precursor for the biosynthesis of both primary aromatic metabolites, including phenylalanine, tyrosine and tryptophan, and secondary aromatic metabolites, including phenazine-1-carboxylic acid (PCA) and pyocyanin (PYO). The enzyme 3-deoxy-d- arabino -heptulosonate 7-phosphate synthase (DAH7PS) catalyses the first committed step of the shikimate pathway, en route to chorismate. P. aeruginosa expresses multiple, distinct DAH7PSs that are associated with either primary or secondary aromatic compound biosynthesis. Here we report the structure of a type II DAH7PS, encoded by phzC as part of the duplicated phenazine biosynthetic cluster, from P. aeruginosa (PAO1) revealing for the first time the structure of a type II DAH7PS involved in secondary metabolism. The omission of the structural elements α 2a and α 2b , relative to other characterised type II DAH7PSs, leads to the formation of an alternative, dimeric, solution-state structure for this type II DAH7PS with an oligomeric interface that has not previously been characterised and that does not facilitate the formation of aromatic amino acid allosteric binding sites. The sequence similarity and, in particular, the common N-terminal extension suggest a common origin for the type II DAH7PSs from P. aeruginosa. The results described in the present study support an expanded classification of the type II DAH7PSs as type II A and type II B based on sequence characteristics, structure and function of the resultant proteins, and on defined physiological roles within primary or secondary metabolism.
Organizational Affiliation:
Biomolecular Interaction Centre and School of Physical and Chemical Sciences, University of Canterbury, Christchurch 8041, New Zealand.