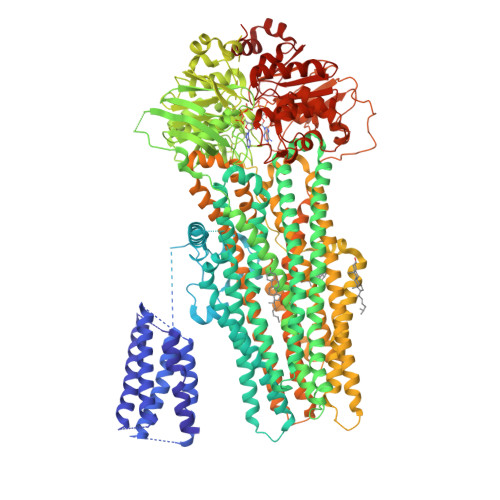
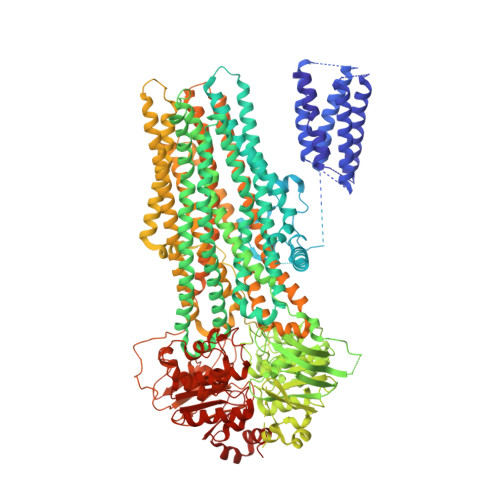
ATP Binding Enables Substrate Release from Multidrug Resistance Protein 1.
Johnson, Z.L., Chen, J.(2018) Cell 172: 81-89.e10
- PubMed: 29290467
- DOI: https://doi.org/10.1016/j.cell.2017.12.005
- Primary Citation of Related Structures:
6BHU - PubMed Abstract:
The multidrug resistance protein MRP1 is an ATP-driven pump that confers resistance to chemotherapy. Previously, we have shown that intracellular substrates are recruited to a bipartite binding site when the transporter rests in an inward-facing conformation. A key question remains: how are high-affinity substrates transferred across the membrane and released outside the cell? Using electron cryomicroscopy, we show here that ATP binding opens the transport pathway to the extracellular space and reconfigures the substrate-binding site such that it relinquishes its affinity for substrate. Thus, substrate is released prior to ATP hydrolysis. With this result, we now have a complete description of the conformational cycle that enables substrate transfer in a eukaryotic ABC exporter.
Organizational Affiliation:
Laboratory of Membrane Biology and Biophysics, The Rockefeller University and Howard Hughes Medical Institute, New York, NY 10065, USA.