Structure-Based Design of Selective Noncovalent CDK12 Inhibitors.
Johannes, J.W., Denz, C.R., Su, N., Wu, A., Impastato, A.C., Mlynarski, S., Varnes, J.G., Prince, D.B., Cidado, J., Gao, N., Haddrick, M., Jones, N.H., Li, S., Li, X., Liu, Y., Nguyen, T.B., O'Connell, N., Rivers, E., Robbins, D.W., Tomlinson, R., Yao, T., Zhu, X., Ferguson, A.D., Lamb, M.L., Manchester, J.I., Guichard, S.(2018) ChemMedChem 13: 231-235
- PubMed: 29266803
- DOI: https://doi.org/10.1002/cmdc.201700695
- Primary Citation of Related Structures:
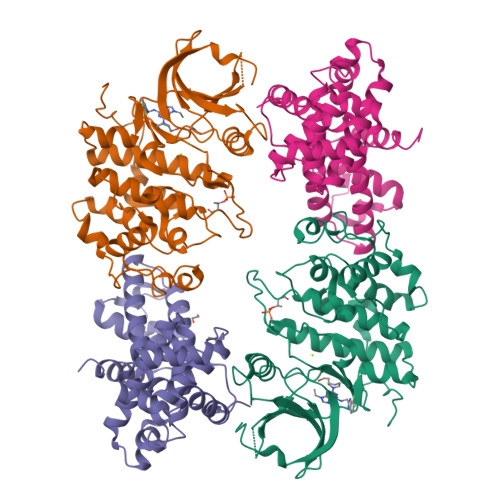
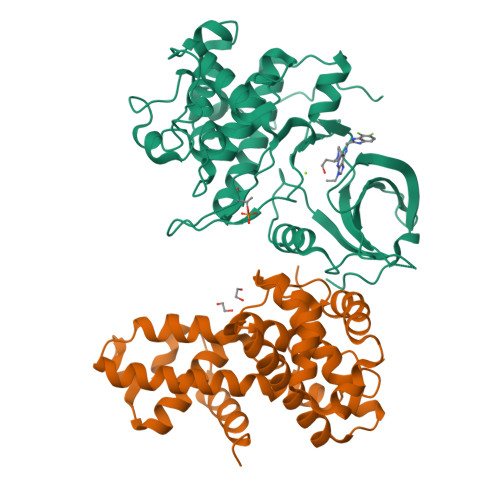
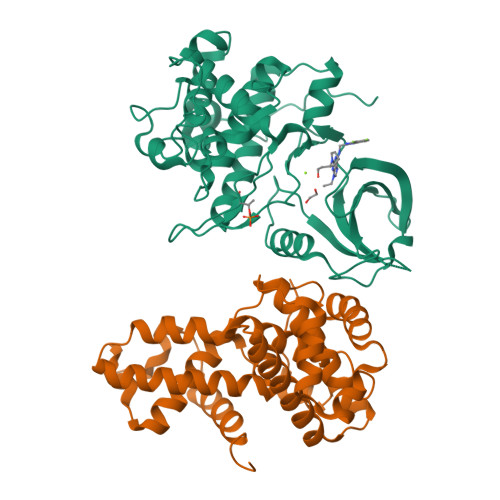
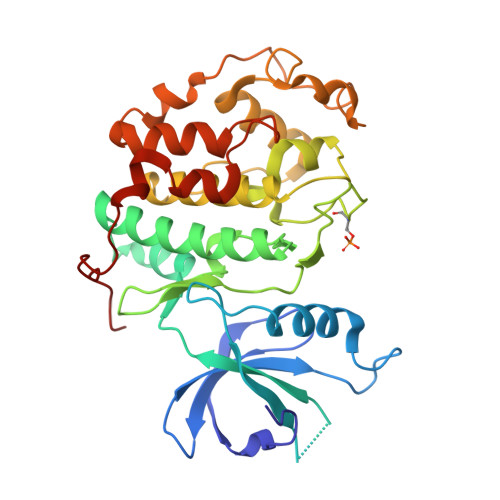
6B3E - PubMed Abstract:
Cyclin-dependent kinase (CDK) 12 knockdown via siRNA decreases the transcription of DNA-damage-response genes and sensitizes BRCA wild-type cells to poly(ADP-ribose) polymerase (PARP) inhibition. To recapitulate this effect with a small molecule, we sought a potent, selective CDK12 inhibitor. Crystal structures and modeling informed hybridization between dinaciclib and SR-3029, resulting in lead compound 5 [(S)-2-(1-(6-(((6,7-difluoro-1H-benzo[d]imidazol-2-yl)methyl)amino)-9-ethyl-9H-purin-2-yl)piperidin-2-yl)ethan-1-ol]. Further structure-guided optimization delivered a series of selective CDK12 inhibitors, including compound 7 [(S)-2-(1-(6-(((6,7-difluoro-1H-benzo[d]imidazol-2-yl)methyl)amino)-9-isopropyl-9H-purin-2-yl)piperidin-2-yl)ethan-1-ol]. Profiling of this compound across CDK9, 7, 2, and 1 at high ATP concentration, single-point kinase panel screening against 352 targets at 0.1 μm, and proteomics via kinase affinity matrix technology demonstrated the selectivity. This series of compounds inhibits phosphorylation of Ser2 on the C-terminal repeat domain of RNA polymerase II, consistent with CDK12 inhibition. These selective compounds were also acutely toxic to OV90 as well as THP1 cells.
Organizational Affiliation:
Oncology, IMED Biotech Unit, AstraZeneca, Boston, MA, USA.