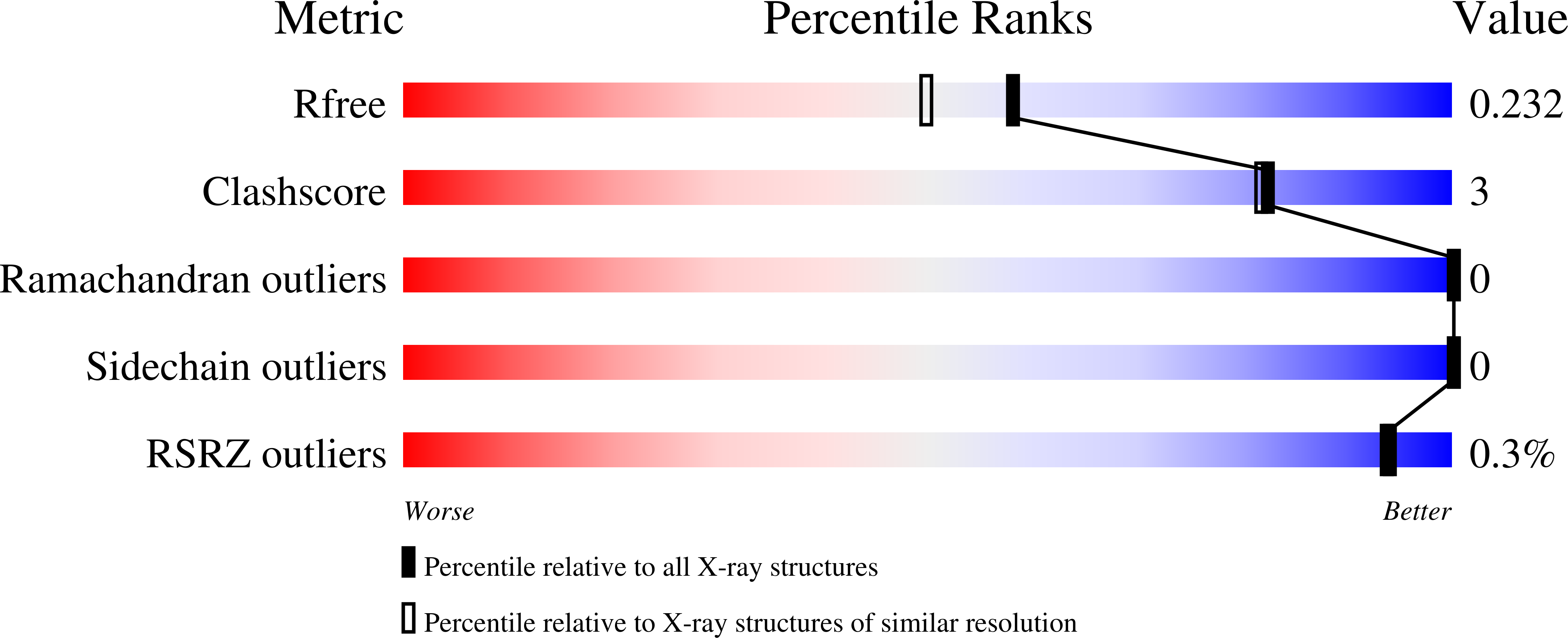
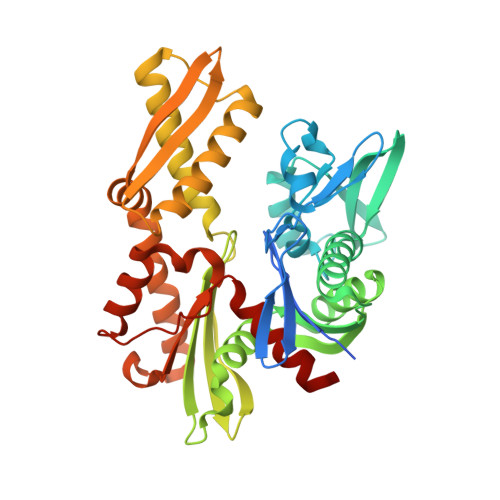
Disrupted Hydrogen-Bond Network and Impaired ATPase Activity in an Hsc70 Cysteine Mutant.
O'Donnell, J.P., Marsh, H.M., Sondermann, H., Sevier, C.S.(2018) Biochemistry 57: 1073-1086
- PubMed: 29300467
- DOI: https://doi.org/10.1021/acs.biochem.7b01005
- Primary Citation of Related Structures:
6B1I, 6B1M, 6B1N - PubMed Abstract:
The ATPase domain of members of the 70 kDa heat shock protein (Hsp70) family shows a high degree of sequence, structural, and functional homology across species. A broadly conserved residue within the Hsp70 ATPase domain that captured our attention is an unpaired cysteine, positioned proximal to the site of nucleotide binding. Prior studies of several Hsp70 family members show this cysteine is not required for Hsp70 ATPase activity, yet select amino acid replacements of the cysteine can dramatically alter ATP hydrolysis. Moreover, post-translational modification of the cysteine has been reported to limit ATP hydrolysis for several Hsp70s. To better understand the underlying mechanism for how perturbation of this noncatalytic residue modulates Hsp70 function, we determined the structure for a cysteine-to-tryptophan mutation in the constitutively expressed, mammalian Hsp70 family member Hsc70. Our work reveals that the steric hindrance produced by a cysteine-to-tryptophan mutation disrupts the hydrogen-bond network within the active site, resulting in a loss of proper catalytic magnesium coordination. We propose that a similarly altered active site is likely observed upon post-translational oxidation. We speculate that the subtle changes we detect in the hydrogen-bonding network may relate to the previously reported observation that cysteine oxidation can influence Hsp70 interdomain communication.
Organizational Affiliation:
Department of Molecular Medicine, Cornell University , Ithaca, New York 14853, United States.