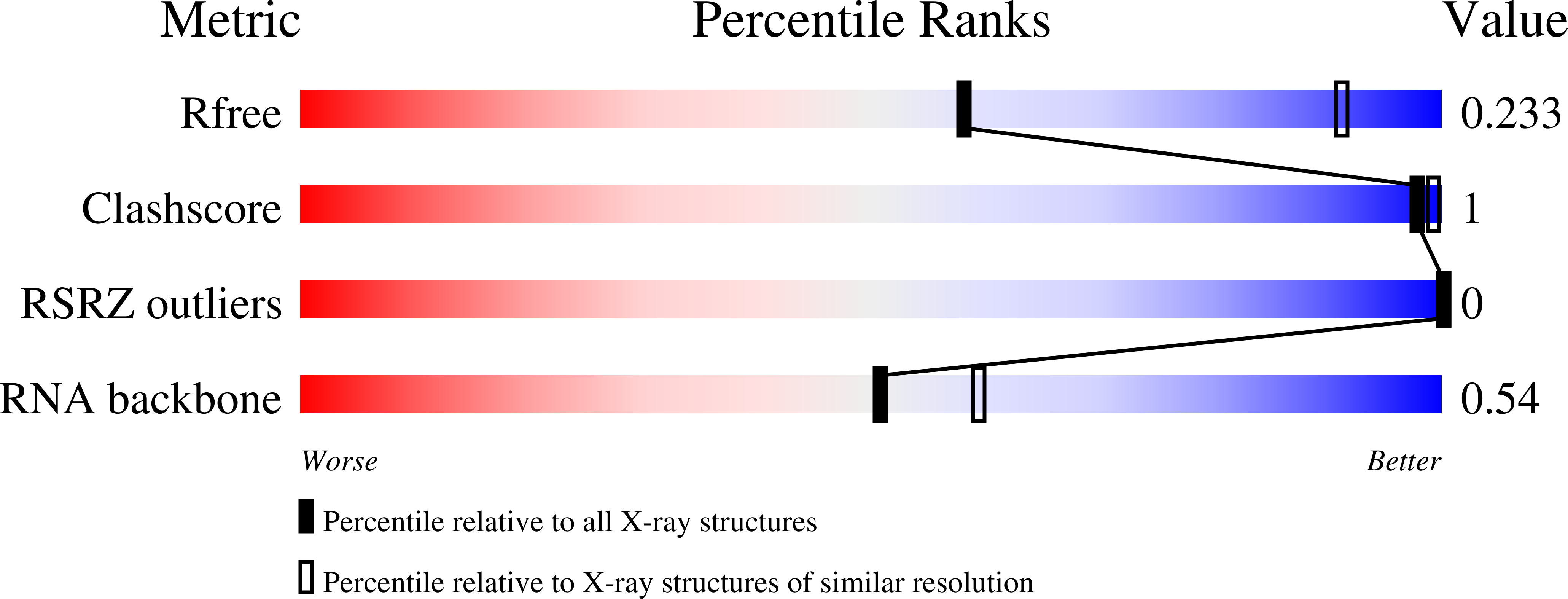
Structural Rationale for the Enhanced Catalysis of Nonenzymatic RNA Primer Extension by a Downstream Oligonucleotide.
Zhang, W., Tam, C.P., Zhou, L., Oh, S.S., Wang, J., Szostak, J.W.(2018) J Am Chem Soc 140: 2829-2840
- PubMed: 29411978
- DOI: https://doi.org/10.1021/jacs.7b11750
- Primary Citation of Related Structures:
5UX3, 5UZ6, 5V0H, 5V0J, 5V0O, 5V9Z, 5VCF, 5VCI, 5VGW, 6AZ4, 6BMD - PubMed Abstract:
Nonenzymatic RNA primer extension by activated mononucleotides has long served as a model for the study of prebiotic RNA copying. We have recently shown that the rate of primer extension is greatly enhanced by the formation of an imidazolium-bridged dinucleotide between the incoming monomer and a second, downstream activated monomer. However, the rate of primer extension is further enhanced if the downstream monomer is replaced by an activated oligonucleotide. Even an unactivated downstream oligonucleotide provides a modest enhancement in the rate of reaction of a primer with a single activated monomer. Here we study the mechanism of these effects through crystallographic studies of RNA complexes with the recently synthesized nonhydrolyzable substrate analog, guanosine 5'-(4-methylimidazolyl)-phosphonate (ICG). ICG mimics 2-methylimidazole activated guanosine-5'-phosphate (2-MeImpG), a commonly used substrate in nonenzymatic primer extension experiments. We present crystal structures of primer-template complexes with either one or two ICG residues bound downstream of a primer. In both cases, the aryl-phosphonate moiety of the ICG adjacent to the primer is disordered. To investigate the effect of a downstream oligonucleotide, we transcribed a short RNA oligonucleotide with either a 5'-ICG residue, a 5'-phosphate or a 5'-hydroxyl. We then determined crystal structures of primer-template complexes with a bound ICG monomer sandwiched between the primer and each of the three downstream oligonucleotides. Surprisingly, all three oligonucleotides rigidify the ICG monomer conformation and position it for attack by the primer 3'-hydroxyl. Furthermore, when GpppG, an analog of the imidazolium-bridged intermediate, is sandwiched between an upstream primer and a downstream helper oligonucleotide, or covalently linked to the 5'-end of the downstream oligonucleotide, the complex is better preorganized for primer extension than in the absence of a downstream oligonucleotide. Our results suggest that a downstream helper oligonucleotide contributes to the catalysis of primer extension by favoring a reactive conformation of the primer-template-intermediate complex.
Organizational Affiliation:
Howard Hughes Medical Institute and Center for Computational and Integrative Biology, Massachusetts General Hospital , Boston, Massachusetts 02114, United States.