Biochemical and Structural Analysis of Substrate Specificity of a Phenylalanine Ammonia-Lyase.
Jun, S.Y., Sattler, S.A., Cortez, G.S., Vermerris, W., Sattler, S.E., Kang, C.(2018) Plant Physiol 176: 1452-1468
- PubMed: 29196539
- DOI: https://doi.org/10.1104/pp.17.01608
- Primary Citation of Related Structures:
6AT7 - PubMed Abstract:
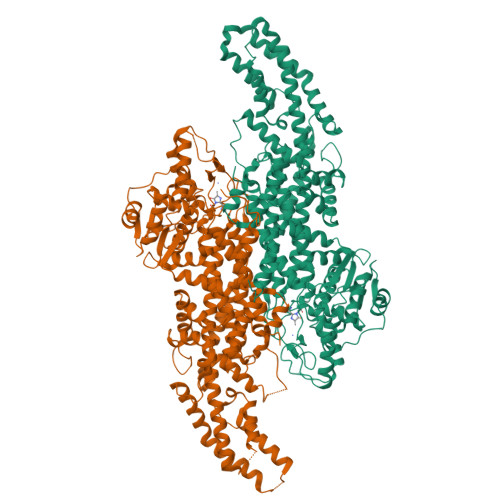
Phenylalanine ammonia-lyase (PAL) is the first enzyme of the general phenylpropanoid pathway catalyzing the nonoxidative elimination of ammonia from l-phenylalanine to give trans -cinnamate. In monocots, PAL also displays tyrosine ammonia lyase (TAL) activity, leading to the formation of p -coumaric acid. The catalytic mechanism and substrate specificity of a major PAL from sorghum ( Sorghum bicolor ; SbPAL1), a strategic plant for bioenergy production, were deduced from crystal structures, molecular docking, site-directed mutagenesis, and kinetic and thermodynamic analyses. This first crystal structure of a monocotyledonous PAL displayed a unique conformation in its flexible inner loop of the 4-methylidene-imidazole-5-one (MIO) domain compared with that of dicotyledonous plants. The side chain of histidine-123 in the MIO domain dictated the distance between the catalytic MIO prosthetic group created from 189 Ala-Ser-Gly 191 residues and the bound l-phenylalanine and l-tyrosine, conferring the deamination reaction through either the Friedel-Crafts or E 2 reaction mechanism. Several recombinant mutant SbPAL1 enzymes were generated via structure-guided mutagenesis, one of which, H123F-SbPAL1, has 6.2 times greater PAL activity without significant TAL activity. Additional PAL isozymes of sorghum were characterized and categorized into three groups. Taken together, this approach identified critical residues and explained substrate preferences among PAL isozymes in sorghum and other monocots, which can serve as the basis for the engineering of plants with enhanced biomass conversion properties, disease resistance, or nutritional quality.
Organizational Affiliation:
Department of Chemistry, Washington State University, Pullman, Washington 99164.