Targeting Allostery with Avatars to Design Inhibitors Assessed by Cell Activity: Dissecting MRE11 Endo- and Exonuclease Activities.
Moiani, D., Ronato, D.A., Brosey, C.A., Arvai, A.S., Syed, A., Masson, J.Y., Petricci, E., Tainer, J.A.(2018) Methods Enzymol 601: 205-241
- PubMed: 29523233
- DOI: https://doi.org/10.1016/bs.mie.2017.11.030
- Primary Citation of Related Structures:
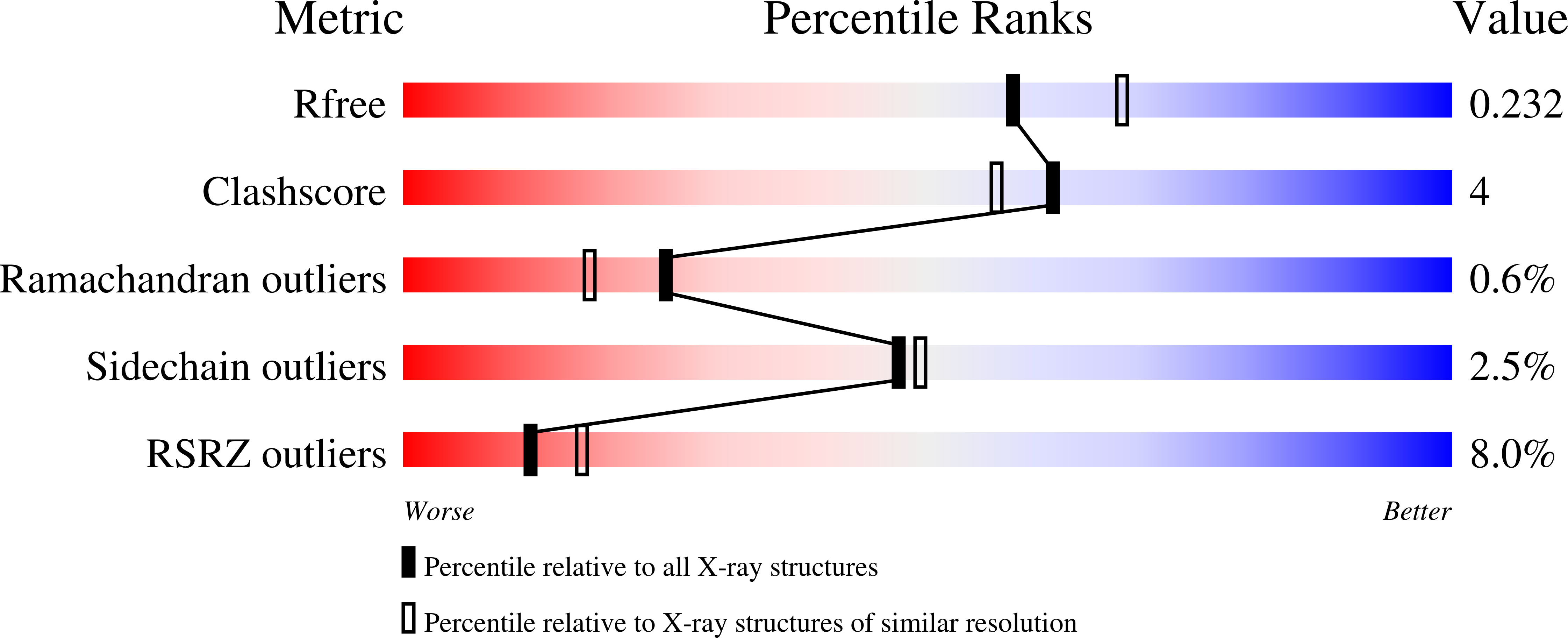
6ASC - PubMed Abstract:
For inhibitor design, as in most research, the best system is question dependent. We suggest structurally defined allostery to design specific inhibitors that target regions beyond active sites. We choose systems allowing efficient quality structures with conformational changes as optimal for structure-based design to optimize inhibitors. We maintain that evolutionarily related targets logically provide molecular avatars, where this Sanskrit term for descent includes ideas of functional relationships and of being a physical embodiment of the target's essential features without requiring high sequence identity. Appropriate biochemical and cell assays provide quantitative measurements, and for biomedical impacts, any inhibitor's activity should be validated in human cells. Specificity is effectively shown empirically by testing if mutations blocking target activity remove cellular inhibitor impact. We propose this approach to be superior to experiments testing for lack of cross-reactivity among possible related enzymes, which is a challenging negative experiment. As an exemplary avatar system for protein and DNA allosteric conformational controls, we focus here on developing separation-of-function inhibitors for meiotic recombination 11 nuclease activities. This was achieved not by targeting the active site but rather by geometrically impacting loop motifs analogously to ribosome antibiotics. These loops are neighboring the dimer interface and active site act in sculpting dsDNA and ssDNA into catalytically competent complexes. One of our design constraints is to preserve DNA substrate binding to geometrically block competing enzymes and pathways from the damaged site. We validate our allosteric approach to controlling outcomes in human cells by reversing the radiation sensitivity and genomic instability in BRCA mutant cells.
Organizational Affiliation:
The University of Texas, M.D. Anderson Cancer Center, Houston, TX, United States.