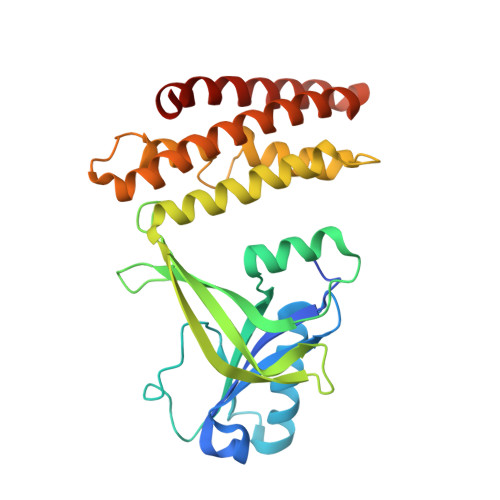
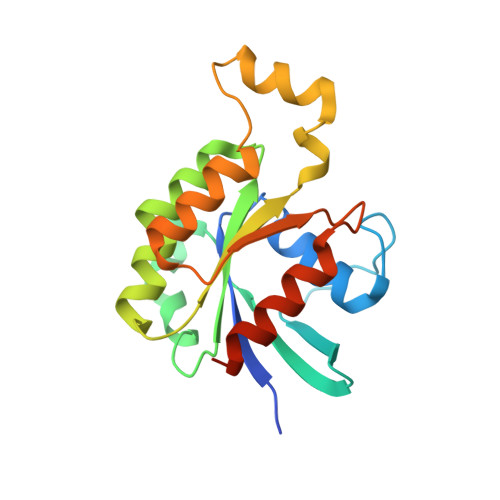
Structural Basis for the Dual Substrate Specificity of DOCK7 Guanine Nucleotide Exchange Factor.
Kukimoto-Niino, M., Tsuda, K., Ihara, K., Mishima-Tsumagari, C., Honda, K., Ohsawa, N., Shirouzu, M.(2019) Structure 27: 741
- PubMed: 30853411
- DOI: https://doi.org/10.1016/j.str.2019.02.001
- Primary Citation of Related Structures:
6AJ4, 6AJL - PubMed Abstract:
The Dedicator Of CytoKinesis (DOCK) family of atypical guanine nucleotide exchange factors activates the Rho family GTPases Rac and/or Cdc42 through DOCK homology region 2 (DHR-2). Previous structural analyses of the DHR-2 domains of DOCK2 and DOCK9 have shown that they preferentially bind Rac1 and Cdc42, respectively; however, the molecular mechanism by which DHR-2 distinguishes between these GTPases is unclear. Here we report the crystal structure of the Cdc42-bound form of the DOCK7 DHR-2 domain showing dual specificity for Rac1 and Cdc42. The structure revealed increased substrate tolerance of DOCK7 at the interfaces with switch 1 and residue 56 of Cdc42. Furthermore, molecular dynamics simulations showed a closed-to-open conformational change in the DOCK7 DHR-2 domain between the Cdc42- and Rac1-bound states by lobe B displacement. Our results suggest that lobe B acts as a sensor for identifying different switch 1 conformations and explain how DOCK7 recognizes both Rac1 and Cdc42.
Organizational Affiliation:
Laboratory for Protein Functional and Structural Biology, RIKEN Center for Biosystems Dynamics Research, Yokohama, Kanagawa 230-0045, Japan; Division of Structural and Synthetic Biology, RIKEN Center for Life Science Technologies, Yokohama, Kanagawa 230-0045, Japan. Electronic address: kukimoto@riken.jp.