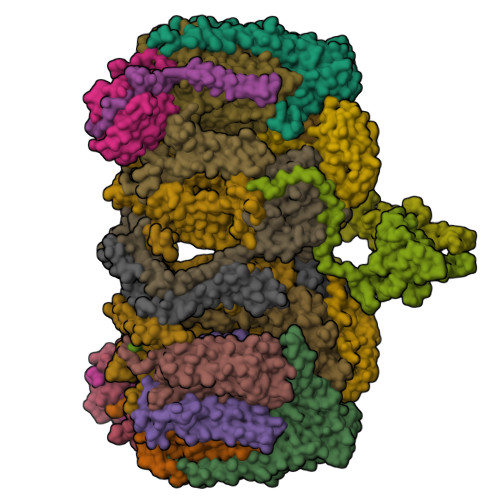
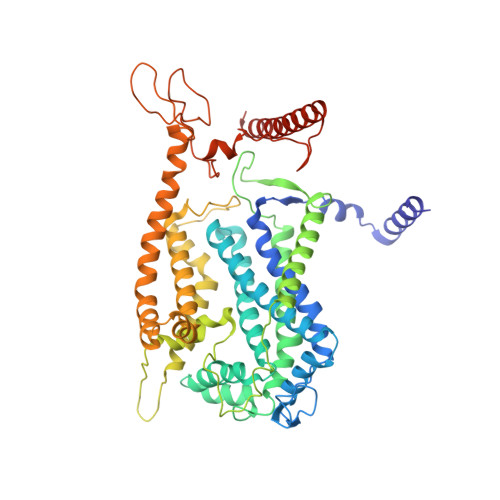
An electron transfer path connects subunits of a mycobacterial respiratory supercomplex.
Gong, H., Li, J., Xu, A., Tang, Y., Ji, W., Gao, R., Wang, S., Yu, L., Tian, C., Li, J., Yen, H.Y., Man Lam, S., Shui, G., Yang, X., Sun, Y., Li, X., Jia, M., Yang, C., Jiang, B., Lou, Z., Robinson, C.V., Wong, L.L., Guddat, L.W., Sun, F., Wang, Q., Rao, Z.(2018) Science 362
- PubMed: 30361386
- DOI: https://doi.org/10.1126/science.aat8923
- Primary Citation of Related Structures:
6ADQ - PubMed Abstract:
We report a 3.5-angstrom-resolution cryo-electron microscopy structure of a respiratory supercomplex isolated from Mycobacterium smegmatis. It comprises a complex III dimer flanked on either side by individual complex IV subunits. Complex III and IV associate so that electrons can be transferred from quinol in complex III to the oxygen reduction center in complex IV by way of a bridging cytochrome subunit. We observed a superoxide dismutase-like subunit at the periplasmic face, which may be responsible for detoxification of superoxide formed by complex III. The structure reveals features of an established drug target and provides a foundation for the development of treatments for human tuberculosis.
Organizational Affiliation:
State Key Laboratory of Medicinal Chemical Biology and College of Life Science, Nankai University, Tianjin 300353, China.