Structural basis for the modulation of voltage-gated sodium channels by animal toxins.
Shen, H., Li, Z., Jiang, Y., Pan, X., Wu, J., Cristofori-Armstrong, B., Smith, J.J., Chin, Y.K.Y., Lei, J., Zhou, Q., King, G.F., Yan, N.(2018) Science 362
- PubMed: 30049784
- DOI: https://doi.org/10.1126/science.aau2596
- Primary Citation of Related Structures:
6A90, 6A91, 6A95 - PubMed Abstract:
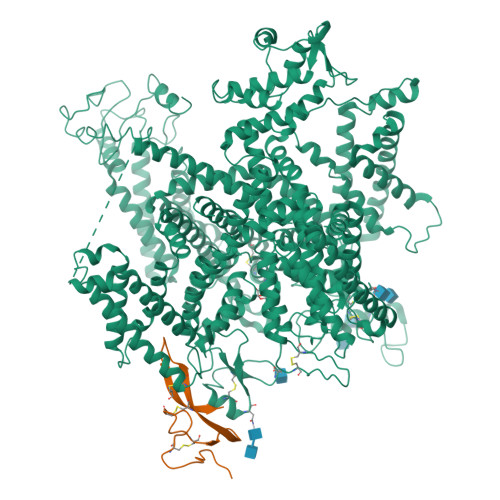
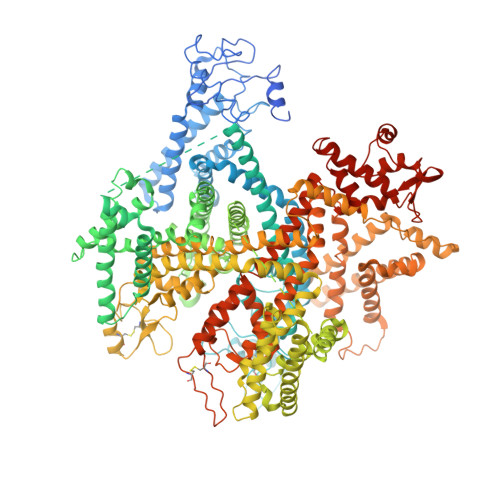
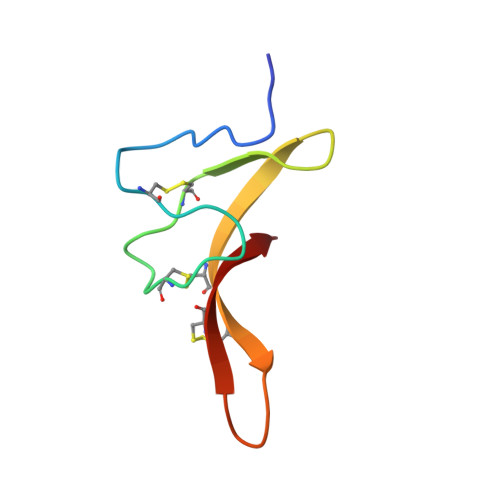
Animal toxins that modulate the activity of voltage-gated sodium (Na v ) channels are broadly divided into two categories-pore blockers and gating modifiers. The pore blockers tetrodotoxin (TTX) and saxitoxin (STX) are responsible for puffer fish and shellfish poisoning in humans, respectively. Here, we present structures of the insect Na v channel Na v PaS bound to a gating modifier toxin Dc1a at 2.8 angstrom-resolution and in the presence of TTX or STX at 2.6-Å and 3.2-Å resolution, respectively. Dc1a inserts into the cleft between VSD II and the pore of Na v PaS, making key contacts with both domains. The structures with bound TTX or STX reveal the molecular details for the specific blockade of Na + access to the selectivity filter from the extracellular side by these guanidinium toxins. The structures shed light on structure-based development of Na v channel drugs.
Organizational Affiliation:
State Key Laboratory of Membrane Biology, School of Life Sciences and School of Medicine, Tsinghua University, Beijing 100084, China.