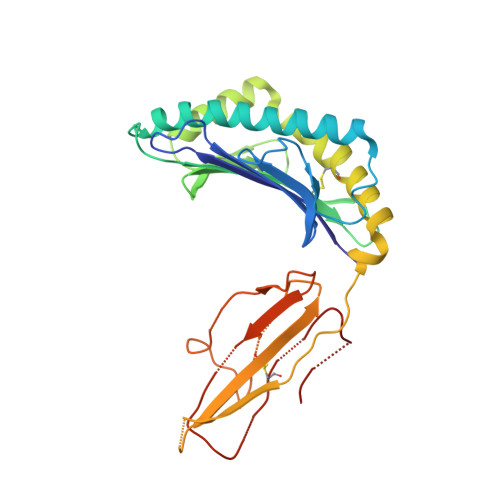
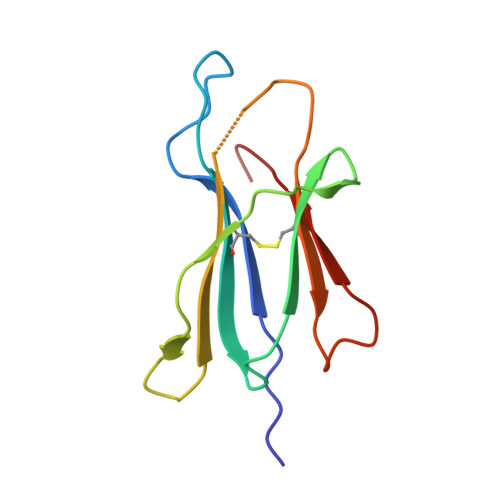
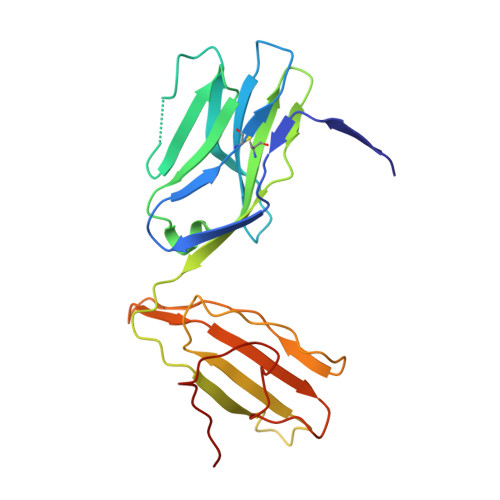
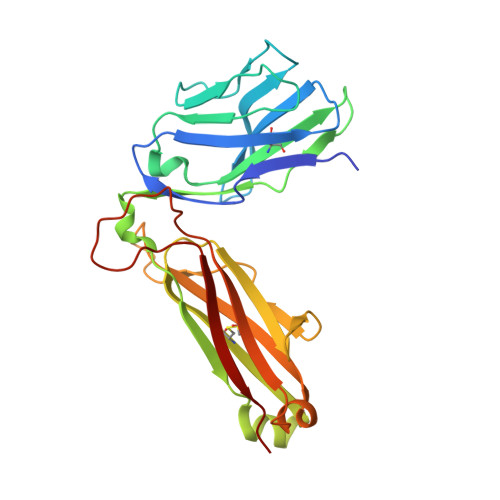
How an alloreactive T-cell receptor achieves peptide and MHC specificity.
Wang, Y., Singh, N.K., Spear, T.T., Hellman, L.M., Piepenbrink, K.H., McMahan, R.H., Rosen, H.R., Vander Kooi, C.W., Nishimura, M.I., Baker, B.M.(2017) Proc Natl Acad Sci U S A 114: E4792-E4801
- PubMed: 28572406
- DOI: https://doi.org/10.1073/pnas.1700459114
- Primary Citation of Related Structures:
5JZI - PubMed Abstract:
T-cell receptor (TCR) allorecognition is often presumed to be relatively nonspecific, attributable to either a TCR focus on exposed major histocompatibility complex (MHC) polymorphisms or the degenerate recognition of allopeptides. However, paradoxically, alloreactivity can proceed with high peptide and MHC specificity. Although the underlying mechanisms remain unclear, the existence of highly specific alloreactive TCRs has led to their use as immunotherapeutics that can circumvent central tolerance and limit graft-versus-host disease. Here, we show how an alloreactive TCR achieves peptide and MHC specificity. The HCV1406 TCR was cloned from T cells that expanded when a hepatitis C virus (HCV)-infected HLA-A2 - individual received an HLA-A2 + liver allograft. HCV1406 was subsequently shown to recognize the HCV nonstructural protein 3 (NS3):1406-1415 epitope with high specificity when presented by HLA-A2. We show that NS3/HLA-A2 recognition by the HCV1406 TCR is critically dependent on features unique to both the allo-MHC and the NS3 epitope. We also find cooperativity between structural mimicry and a crucial peptide "hot spot" and demonstrate its role, along with the MHC, in directing the specificity of allorecognition. Our results help explain the paradox of specificity in alloreactive TCRs and have implications for their use in immunotherapy and related efforts to manipulate TCR recognition, as well as alloreactivity in general.
- Department of Chemistry and Biochemistry, University of Notre Dame, Notre Dame, IN 46556.
Organizational Affiliation: