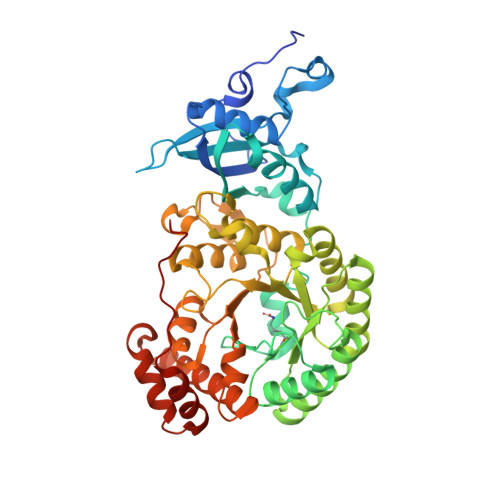
Structure of Rubisco from Arabidopsis thaliana in complex with 2-carboxyarabinitol-1,5-bisphosphate.
Valegard, K., Hasse, D., Andersson, I., Gunn, L.H.(2018) Acta Crystallogr D Struct Biol 74: 1-9
- PubMed: 29372894
- DOI: https://doi.org/10.1107/S2059798317017132
- Primary Citation of Related Structures:
5IU0 - PubMed Abstract:
The crystal structure of ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) from Arabidopsis thaliana is reported at 1.5 Å resolution. In light of the importance of A. thaliana as a model organism for understanding higher plant biology, and the pivotal role of Rubisco in photosynthetic carbon assimilation, there has been a notable absence of an A. thaliana Rubisco crystal structure. A. thaliana Rubisco is an L 8 S 8 hexadecamer comprising eight plastome-encoded catalytic large (L) subunits and eight nuclear-encoded small (S) subunits. A. thaliana produces four distinct small-subunit isoforms (RbcS1A, RbcS1B, RbcS2B and RbcS3B), and this crystal structure provides a snapshot of A. thaliana Rubisco containing the low-abundance RbcS3B small-subunit isoform. Crystals were obtained in the presence of the transition-state analogue 2-carboxy-D-arabinitol-1,5-bisphosphate. A. thaliana Rubisco shares the overall fold characteristic of higher plant Rubiscos, but exhibits an interesting disparity between sequence and structural relatedness to other Rubisco isoforms. These results provide the structural framework to understand A. thaliana Rubisco and the potential catalytic differences that could be conferred by alternative A. thaliana Rubisco small-subunit isoforms.
- Laboratory of Molecular Biophysics, Department of Cell and Molecular Biology, Uppsala University, Husargatan 3, Box 596, SE-751 24 Uppsala, Sweden.
Organizational Affiliation: