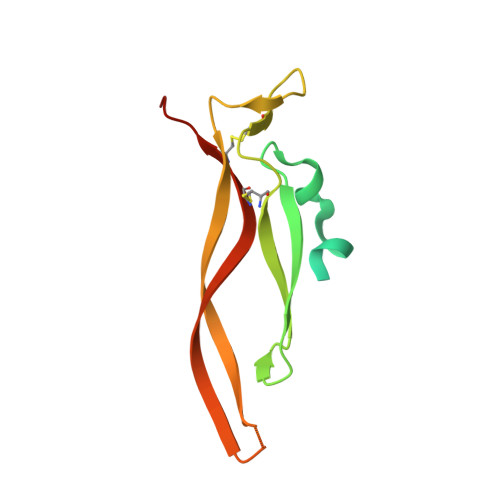
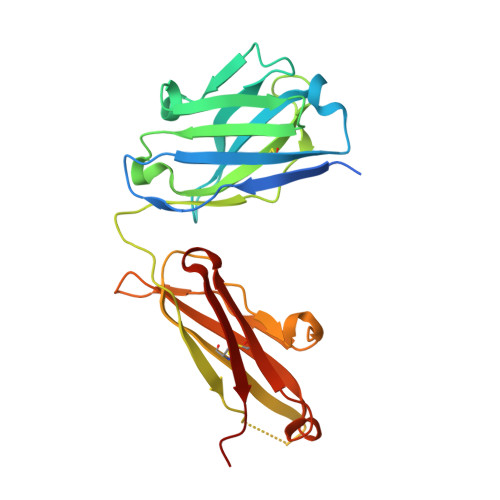
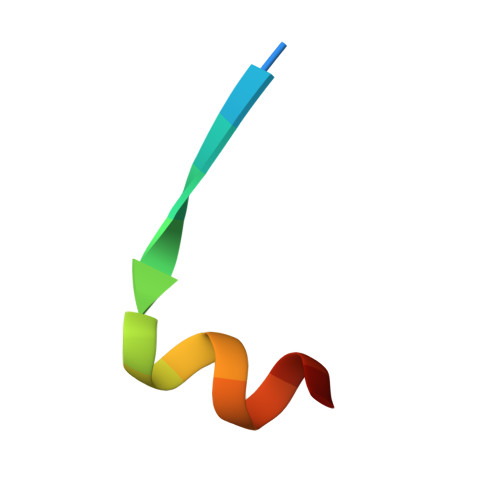
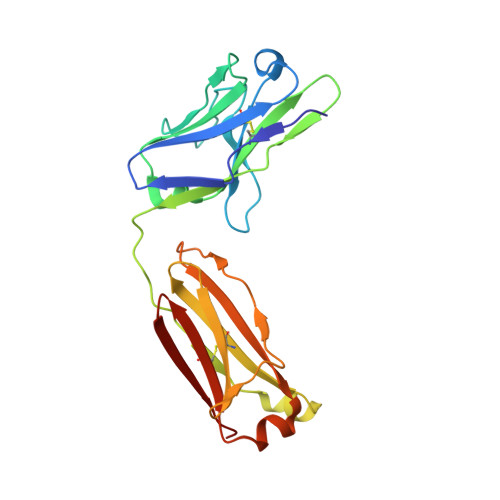
Inhibiting complex IL-17A and IL-17RA interactions with a linear peptide.
Liu, S., Desharnais, J., Sahasrabudhe, P.V., Jin, P., Li, W., Oates, B.D., Shanker, S., Banker, M.E., Chrunyk, B.A., Song, X., Feng, X., Griffor, M., Jimenez, J., Chen, G., Tumelty, D., Bhat, A., Bradshaw, C.W., Woodnutt, G., Lappe, R.W., Thorarensen, A., Qiu, X., Withka, J.M., Wood, L.D.(2016) Sci Rep 6: 26071-26071
- PubMed: 27184415
- DOI: https://doi.org/10.1038/srep26071
- Primary Citation of Related Structures:
5HHV, 5HHX - PubMed Abstract:
IL-17A is a pro-inflammatory cytokine that has been implicated in autoimmune and inflammatory diseases. Monoclonal antibodies inhibiting IL-17A signaling have demonstrated remarkable efficacy, but an oral therapy is still lacking. A high affinity IL-17A peptide antagonist (HAP) of 15 residues was identified through phage-display screening followed by saturation mutagenesis optimization and amino acid substitutions. HAP binds specifically to IL-17A and inhibits the interaction of the cytokine with its receptor, IL-17RA. Tested in primary human cells, HAP blocked the production of multiple inflammatory cytokines. Crystal structure studies revealed that two HAP molecules bind to one IL-17A dimer symmetrically. The N-terminal portions of HAP form a β-strand that inserts between two IL-17A monomers while the C-terminal section forms an α helix that directly blocks IL-17RA from binding to the same region of IL-17A. This mode of inhibition suggests opportunities for developing peptide antagonists against this challenging target.
- Worldwide Research and Development, Pfizer Inc., Eastern Point Road, Groton, CT 06340 USA.
Organizational Affiliation: