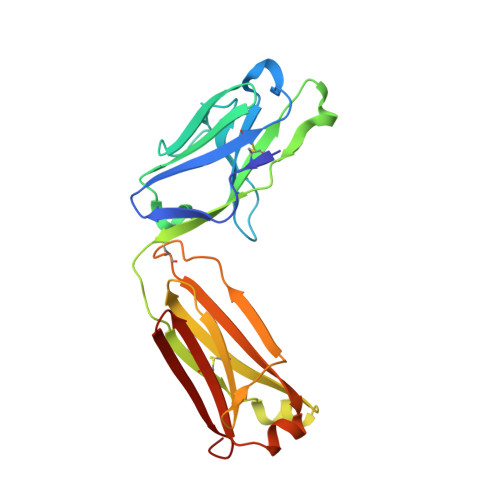
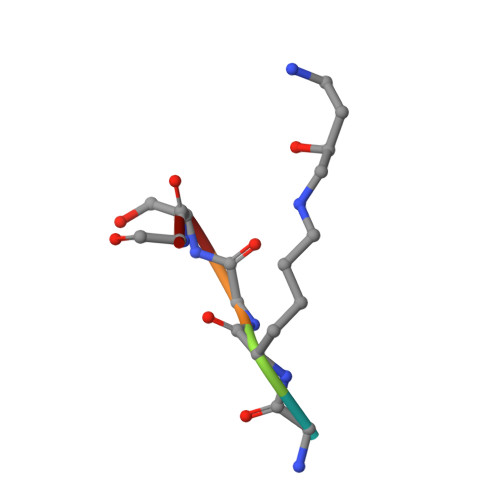
Structural Analysis and Optimization of Context-Independent Anti-Hypusine Antibodies.
Zhai, Q., He, M., Song, A., Deshayes, K., Dixit, V.M., Carter, P.J.(2016) J Mol Biology 428: 603-617
- PubMed: 26778617
- DOI: https://doi.org/10.1016/j.jmb.2016.01.006
- Primary Citation of Related Structures:
5DRN, 5DS8, 5DSC, 5DTF, 5DUB - PubMed Abstract:
Context-independent anti-hypusine antibodies that bind to the post-translational modification (PTM), hypusine, with minimal dependence on flanking amino acid sequences, were identified. The antibodies bind to both hypusine and deoxyhypusine or selectively to hypusine but not to deoxyhypusine. Phage display was used to further enhance the affinity of the antibodies. Affinity maturation of these anti-hypusine antibodies improved their performance in affinity capture of the only currently known hypusinated protein, eukaryotic translation initiation factor 5A. These anti-hypusine antibodies may have utility in the identification of novel hypusinated proteins. Crystal structures of the corresponding Fab fragments were determined in complex with hypusine- or deoxyhypusine-containing peptides. The hypusine or deoxyhypusine moiety was found to reside in a deep pocket formed between VH and VL domains of the Fab fragments. Interaction between the antibodies and hypusine includes an extensive hydrogen bond network. These are, to our knowledge, the first reported structures of context-independent anti-PTM antibodies in complex with the corresponding PTM.
- Department of Antibody Engineering, Genentech Inc., 1 DNA Way, South San Francisco, CA 94080, USA.
Organizational Affiliation: