Identification of the Histidine Residue in Vitamin D Receptor That Covalently Binds to Electrophilic Ligands
Yoshizawa, M., Itoh, T., Hori, T., Kato, A., Anami, Y., Yoshimoto, N., Yamamoto, K.(2018) J Med Chem 61: 6339-6349
- PubMed: 29936834
- DOI: https://doi.org/10.1021/acs.jmedchem.8b00774
- Primary Citation of Related Structures:
5ZWE, 5ZWF, 5ZWH, 5ZWI - PubMed Abstract:
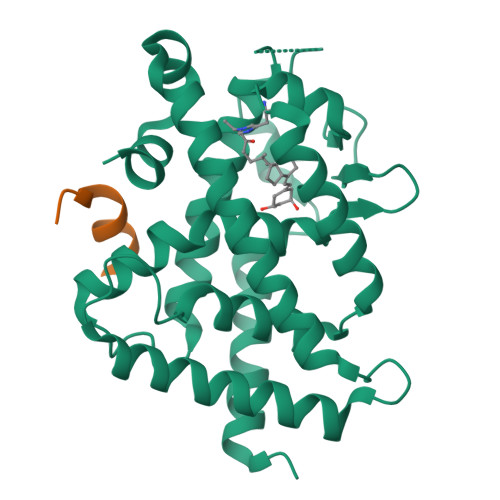
We designed and synthesized vitamin D analogues with an electrophile as covalent modifiers for the vitamin D receptor (VDR). Novel vitamin D analogues 1-4 have an electrophilic enone group at the side chain for conjugate addition to His301 or His393 in the VDR. All compounds showed specific VDR-binding potency and agonistic activity. Covalent bond formations of 1-4 with the ligand-binding domain (LBD) of VDR were evaluated by electrospray ionization mass spectrometry. All compounds were shown to covalently bind to the VDR-LBD, and the abundance of VDR-LBD corresponding conjugate adducts of 1-4 increased with incubation time. Enone compounds 1 and 2 showed higher reactivity than the ene-ynone 3 and dienone 4 compounds. Furthermore, we successfully obtained cocrystals of VDR-LBD with analogues 1-4. X-ray crystallographic analysis showed a covalent bond with His301 in VDR-LBD. We successfully synthesized vitamin D analogues that form a covalent bond with VDR-LBD.
Organizational Affiliation:
Laboratory of Drug Design and Medicinal Chemistry , Showa Pharmaceutical University , 3-3165 Higashi-Tamagawagakuen , Machida, Tokyo 194-8543 , Japan.