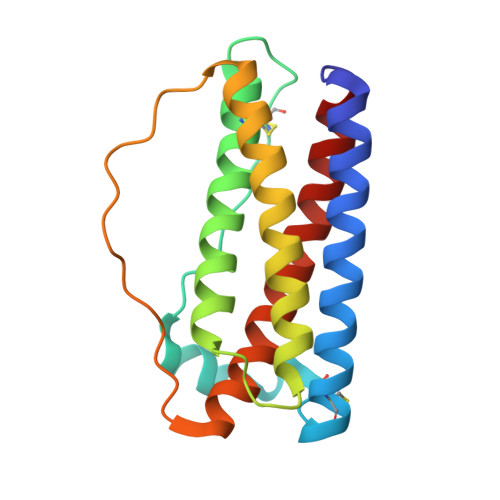
Stabilization of backbone-circularized protein is attained by synergistic gains in enthalpy of folded structure and entropy of unfolded structure.
Shibuya, R., Miyafusa, T., Honda, S.(2019) FEBS J
- PubMed: 31605655
- DOI: https://doi.org/10.1111/febs.15092
- Primary Citation of Related Structures:
5ZO6 - PubMed Abstract:
Backbone circularization is an effective technique for protein stabilization. Here, we investigated the effect of a connector, an engineered segment that connects two protein termini, on the conformational stability of previously designed circularized variants of granulocyte colony-stimulating factor (G-CSF). Heat tolerance and chemical denaturation analyses revealed that aggregation resistance and thermodynamic stability of the circularized variants were superior to those of linear G-CSF. Crystal structure and molecular dynamics (MD) simulation of the most thermodynamically stable variant (C166) revealed a high number of intramolecular hydrogen bonds in both the connector region and Helix D adjacent to the connector region in the folded structure. MD simulations and theoretical calculations involving different force fields indicated a reduction in the main chain entropy of C166 in the unfolded state and increase in the intramolecular hydrogen bond energy of C166 in the folded structure. Although backbone circularization is usually considered to alter chain entropy of the unfolded state, the data indicated that it could also improve the conformational enthalpy of the folded state. Further structural examination of the connector region confirmed that protein design based on a statistical analysis of local structures is an effective approach for predicting an optimum connector length to improve the conformational stability of backbone-circularized proteins. Protein design using backbone circularization with an optimum connector length will be useful for the development of effective and safe protein therapeutics. DATABASE: Structural data are available in Protein Data Bank under the accession number 5ZO6.
Organizational Affiliation:
Department of Computational Biology and Medical Sciences, Graduate School of Frontier Sciences, The University of Tokyo, Kashiwa, Chiba, Japan.