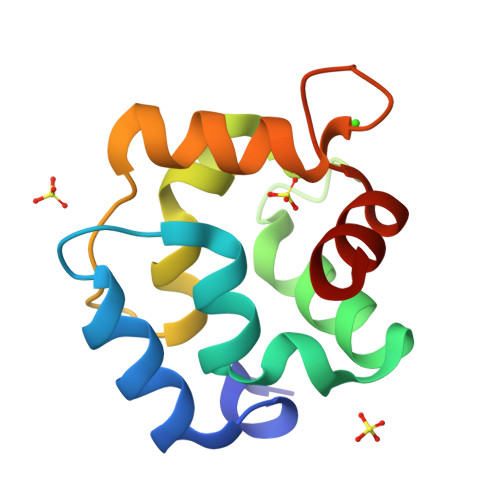
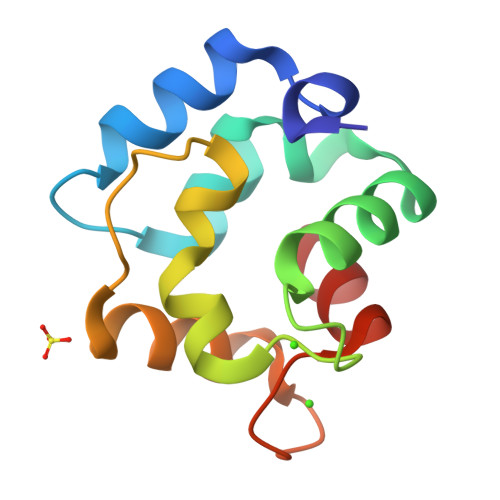
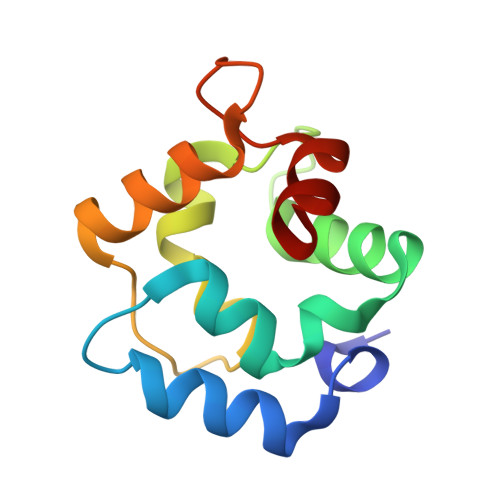
Purification, Characterization, and Crystal Structure of Parvalbumins, the Major Allergens in Mustelus griseus.
Yang, R.Q., Chen, Y.L., Chen, F., Wang, H., Zhang, Q., Liu, G.M., Jin, T., Cao, M.J.(2018) J Agric Food Chem 66: 8150-8159
- PubMed: 29969026
- DOI: https://doi.org/10.1021/acs.jafc.8b01889
- Primary Citation of Related Structures:
5ZGM, 5ZH6 - PubMed Abstract:
Fish play important roles in human nutrition and health, but also trigger allergic reactions in some population. Parvalbumin (PV) represents the major allergen of fish. While IgE cross-reactivity to PV in various bony fish species has been well characterized, little information is available about allergens in cartilaginous fish. In this study, two shark PV isoforms (named as SPV-I and SPV-II) from Mustelus griseus were purified. Their identities were further confirmed by mass spectroscopic analysis. IgE immunoblot analysis showed that sera from fish-allergic patients reacted to both SPV-I and SPV-II, but the majority of sera reacted more intensely to SPV-I than SPV-II. Thermal denaturation monitored by CD spectrum showed that both of the SPV allergens are highly thermostable. SPV-I maintained its IgE-binding capability after heat denaturation, while the IgE-binding capability of SPV-II was reduced. The results of crystal structure showed that SPV-I and SPV-II were similar in their overall tertiary structure, but their amino acid sequences shared lower similarities, indicating that the differences in the IgE-binding capabilities of SPV-I and SPV-II might be due to differential antigen epitopes in these two isoforms.
Organizational Affiliation:
College of Food and Biological Engineering , Jimei University , Xiamen , Fujian 361021 , China.