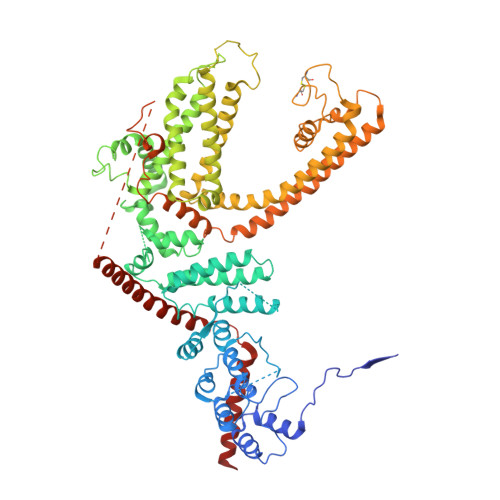
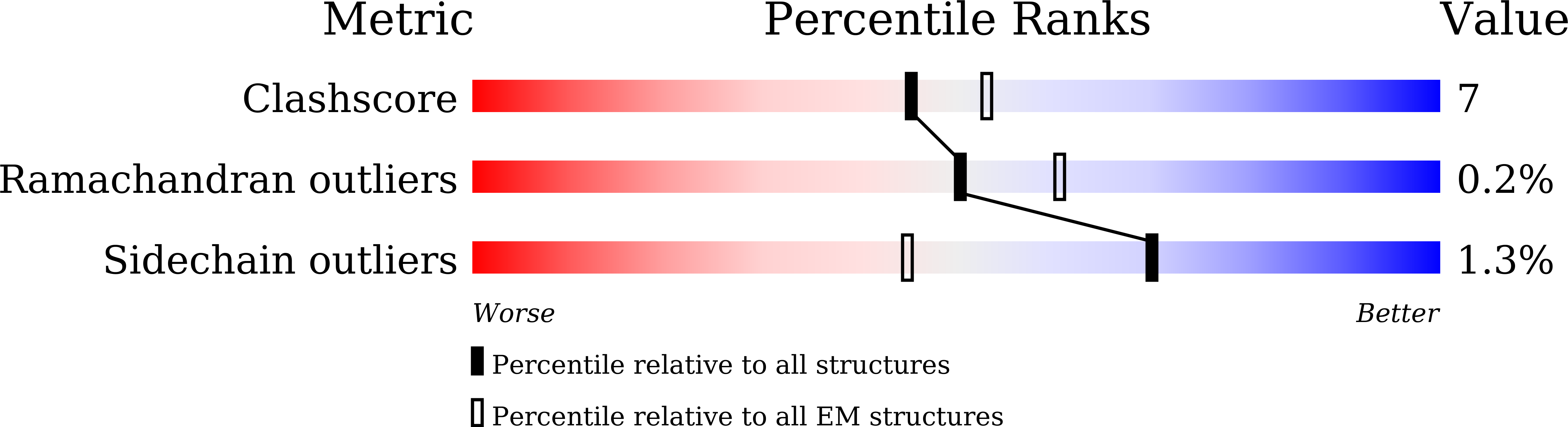Structure of the mouse TRPC4 ion channel.
Duan, J., Li, J., Zeng, B., Chen, G.L., Peng, X., Zhang, Y., Wang, J., Clapham, D.E., Li, Z., Zhang, J.(2018) Nat Commun 9: 3102-3102
- PubMed: 30082700
- DOI: https://doi.org/10.1038/s41467-018-05247-9
- Primary Citation of Related Structures:
5Z96 - PubMed Abstract:
Members of the transient receptor potential (TRP) ion channels conduct cations into cells. They mediate functions ranging from neuronally mediated hot and cold sensation to intracellular organellar and primary ciliary signaling. Here we report a cryo-electron microscopy (cryo-EM) structure of TRPC4 in its unliganded (apo) state to an overall resolution of 3.3 Å. The structure reveals a unique architecture with a long pore loop stabilized by a disulfide bond. Beyond the shared tetrameric six-transmembrane fold, the TRPC4 structure deviates from other TRP channels with a unique cytosolic domain. This unique cytosolic N-terminal domain forms extensive aromatic contacts with the TRP and the C-terminal domains. The comparison of our structure with other known TRP structures provides molecular insights into TRPC4 ion selectivity and extends our knowledge of the diversity and evolution of the TRP channels.
Organizational Affiliation:
School of Basic Medical Sciences, Nanchang University, 330031, Nanchang, Jiangxi, China.