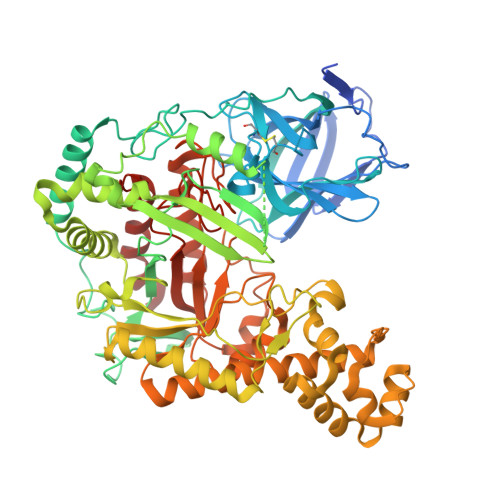
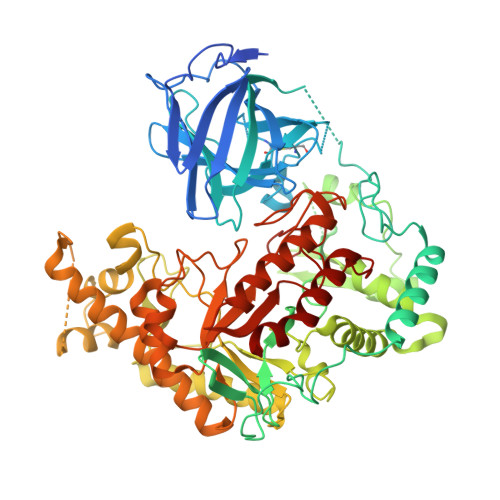
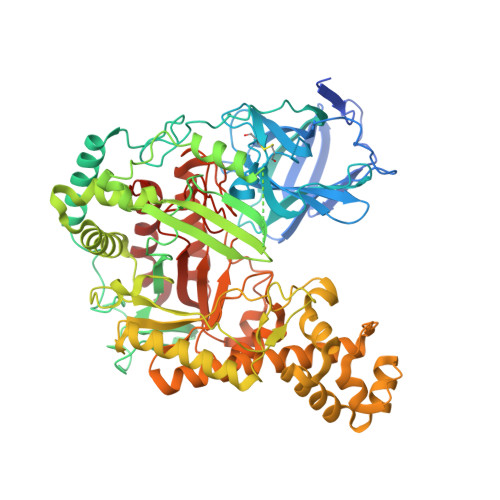
Structure-based design of agarase AgWH50C from Agarivorans gilvus WH0801 to enhance thermostability.
Zhang, P., Zhang, J., Zhang, L., Sun, J., Li, Y., Wu, L., Zhou, J., Xue, C., Mao, X.(2019) Appl Microbiol Biotechnol 103: 1289-1298
- PubMed: 30523371
- DOI: https://doi.org/10.1007/s00253-018-9540-1
- Primary Citation of Related Structures:
5Z6P - PubMed Abstract:
AgWH50C, an exo-β-agarase of GH50 isolated from Agarivorans gilvus WH0801, plays a key role in the enzymatic production of neoagarobiose, which has great application prospect in the cosmetics and pharmaceutical industry. In contrast, the poor thermostability becomes the main obstructive factor of glycoside hydrolase (GH) family 50 agarases, including AgWH50C. Herein, based on the AgWH50C crystal structure, we designed several mutants by a multiple cross-linked rational design protocol used thermostability predicting softwares ETSS, PoPMuSiC, and HotMuSiC. To our surprise, the mutant K621F increased its relative activity by as much as 45% and the optimal temperature increased to 38 °C compared to that of wild-type, AgWH50C (30 °C). The thermostability of K621F also exhibited a substantial improvement. Considering that the gelling temperature of the agarose is higher than 35 °C, K621F can be used to hydrolyze agarose for neoagarobiose production.
Organizational Affiliation:
College of Food Science and Engineering, Ocean University of China, Qingdao, 266003, China.