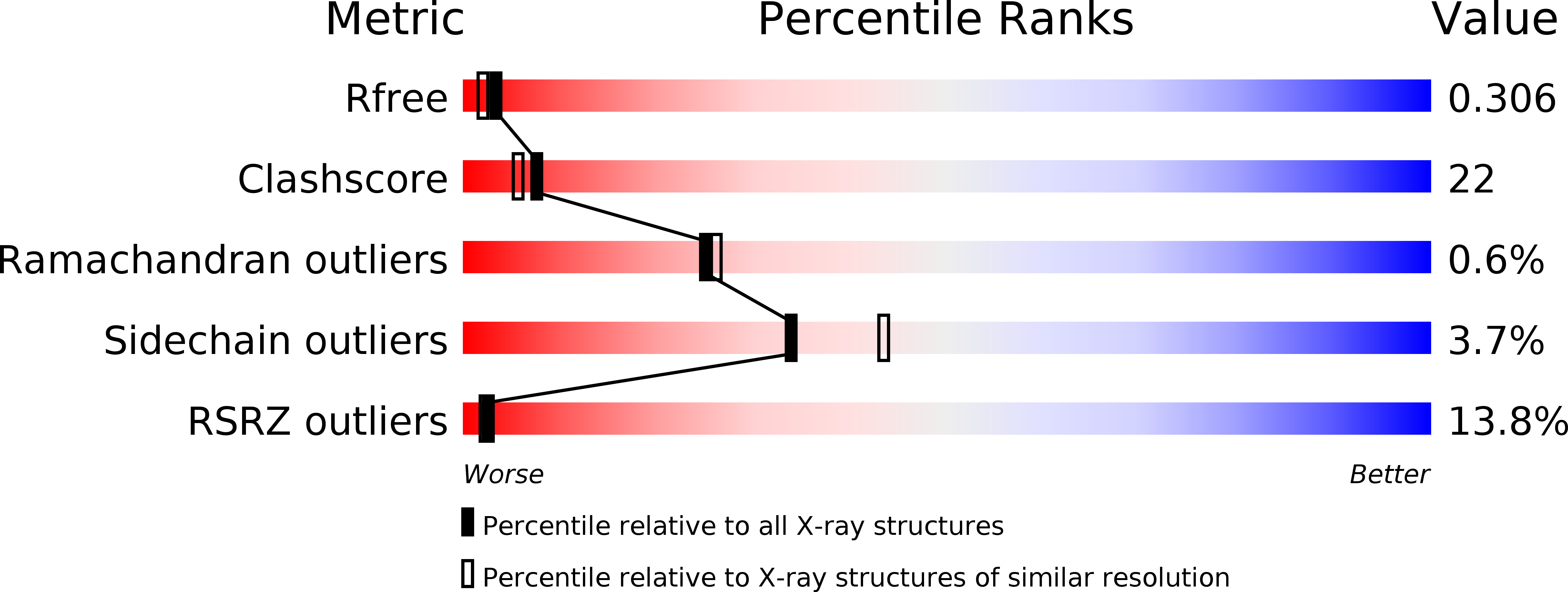
High-pressure protein crystal structure analysis of Escherichia coli dihydrofolate reductase complexed with folate and NADP.
Nagae, T., Yamada, H., Watanabe, N.(2018) Acta Crystallogr D Struct Biol 74: 895-905
- PubMed: 30198899
- DOI: https://doi.org/10.1107/S2059798318009397
- Primary Citation of Related Structures:
4X5F, 4X5G, 4X5H, 4X5I, 4X5J, 5Z6F, 5Z6J, 5Z6K, 5Z6L, 5Z6M - PubMed Abstract:
A high-pressure crystallographic study was conducted on Escherichia coli dihydrofolate reductase (ecDHFR) complexed with folate and NADP + in crystal forms containing both the open and closed conformations of the M20 loop under high-pressure conditions of up to 800 MPa. At pressures between 270 and 500 MPa the crystal form containing the open conformation exhibited a phase transition from P2 1 to C2. Several structural changes in ecDHFR were observed at high pressure that were also accompanied by structural changes in the NADP + cofactor and the hydration structure. In the crystal form with the closed conformation the M20 loop moved as the pressure changed, with accompanying conformational changes around the active site, including NADP + and folate. These movements were consistent with the suggested hypothesis that movement of the M20 loop was necessary for ecDHFR to catalyze the reaction. In the crystal form with the open conformation the nicotinamide ring of the NADP + cofactor undergoes a large flip as an intermediate step in the reaction, despite being in a crystalline state. Furthermore, observation of the water molecules between Arg57 and folate elucidated an early step in the substrate-binding pathway. These results demonstrate the possibility of using high-pressure protein crystallography as a method to capture high-energy substates or transient structures related to the protein reaction cycle.
Organizational Affiliation:
Synchrotron Radiation Research Center, Nagoya University, Chikusa, Nagoya 464-8603, Japan.