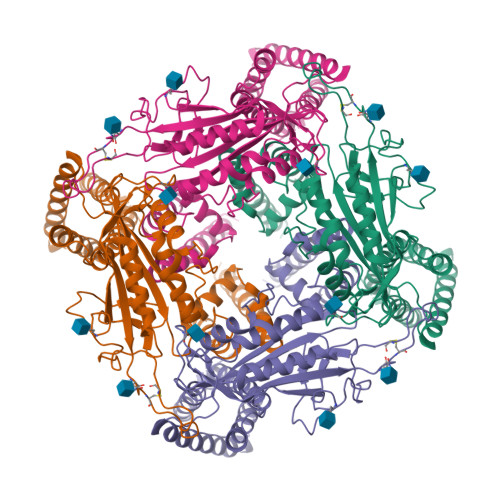
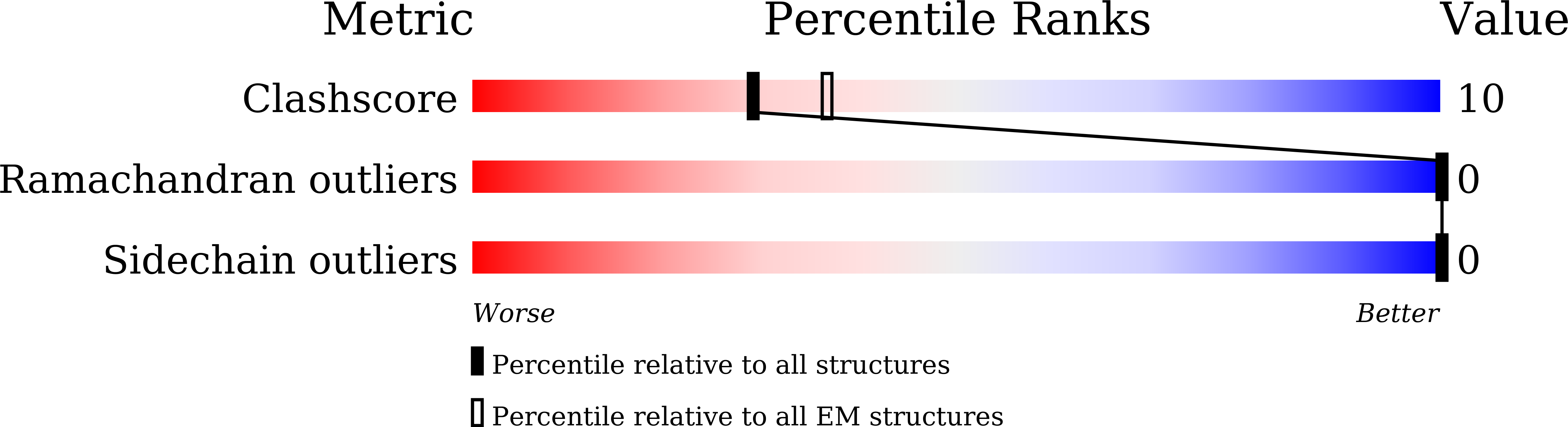Cryo-EM structure of the polycystic kidney disease-like channel PKD2L1.
Su, Q., Hu, F., Liu, Y., Ge, X., Mei, C., Yu, S., Shen, A., Zhou, Q., Yan, C., Lei, J., Zhang, Y., Liu, X., Wang, T.(2018) Nat Commun 9: 1192-1192
- PubMed: 29567962
- DOI: https://doi.org/10.1038/s41467-018-03606-0
- Primary Citation of Related Structures:
5Z1W - PubMed Abstract:
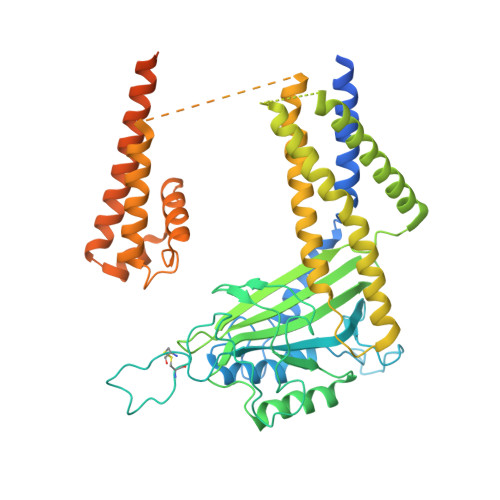
PKD2L1, also termed TRPP3 from the TRPP subfamily (polycystic TRP channels), is involved in the sour sensation and other pH-dependent processes. PKD2L1 is believed to be a nonselective cation channel that can be regulated by voltage, protons, and calcium. Despite its considerable importance, the molecular mechanisms underlying PKD2L1 regulations are largely unknown. Here, we determine the PKD2L1 atomic structure at 3.38 Å resolution by cryo-electron microscopy, whereby side chains of nearly all residues are assigned. Unlike its ortholog PKD2, the pore helix (PH) and transmembrane segment 6 (S6) of PKD2L1, which are involved in upper and lower-gate opening, adopt an open conformation. Structural comparisons of PKD2L1 with a PKD2-based homologous model indicate that the pore domain dilation is coupled to conformational changes of voltage-sensing domains (VSDs) via a series of π-π interactions, suggesting a potential PKD2L1 gating mechanism.
Organizational Affiliation:
Ministry of Education Key Laboratory of Protein Science, Tsinghua University, Beijing, 100084, China.