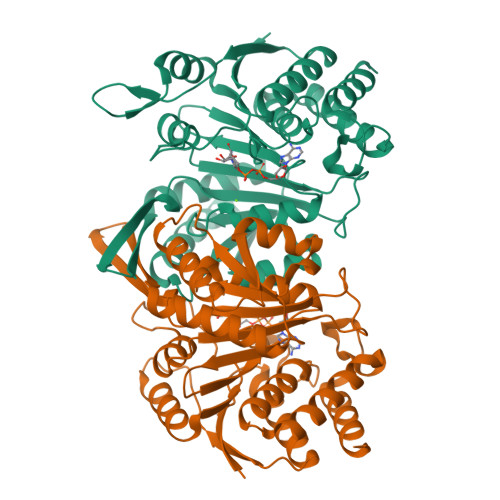
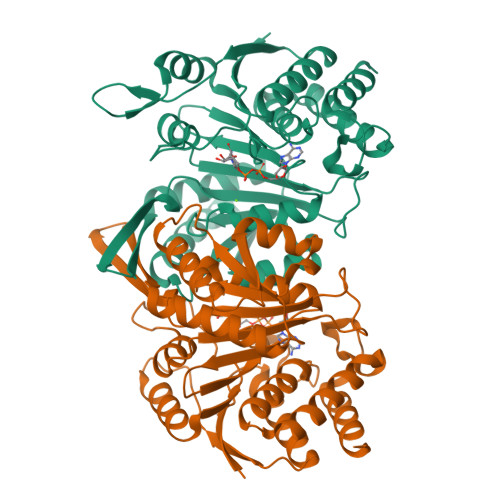
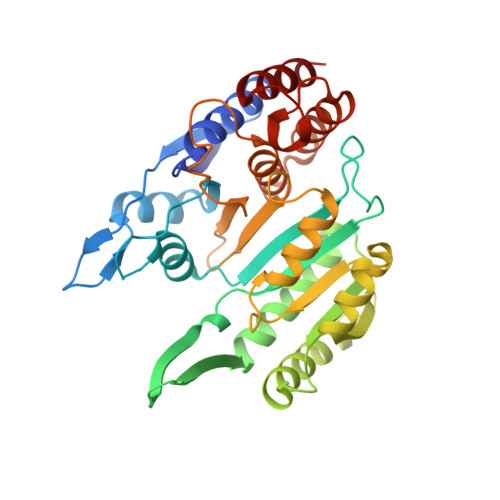
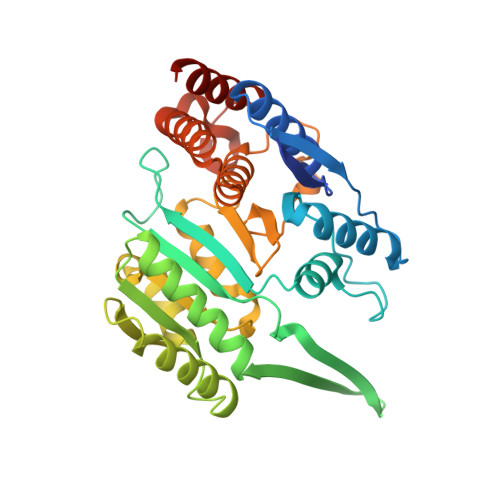
Insights into the inhibitory mechanisms of NADH on the alpha gamma heterodimer of human NAD-dependent isocitrate dehydrogenase.
Liu, Y., Hu, L., Ma, T., Yang, J., Ding, J.(2018) Sci Rep 8: 3146-3146
- PubMed: 29453450
- DOI: https://doi.org/10.1038/s41598-018-21584-7
- Primary Citation of Related Structures:
5YVT - PubMed Abstract:
Human NAD-dependent isocitrate dehydrogenase (NAD-IDH) catalyzes the oxidative decarboxylation of isocitrate in the citric acid cycle. In the α 2 βγ heterotetramer of NAD-IDH, the γ subunit plays the regulatory role and the β subunit the structural role. Previous biochemical data have shown that mammalian NAD-IDHs can be inhibited by NADH; however, the molecular mechanism is unclear. In this work, we show that the αβ, αγ and α 2 βγ enzymes of human NAD-IDH can be inhibited by NADH, and further determine the crystal structure of the αγ heterodimer bound with an Mg 2+ and an NADH at the active site and an NADH at the allosteric site, which resembles that of the inactive α Mg γ heterodimer. The NADH at the active site occupies the binding site for NAD + and prevents the binding of the cofactor. The NADH at the allosteric site occupies the binding sites for ADP and citrate and blocks the binding of the activators. The biochemical data confirm that the NADH binding competes with the binding of NAD + and the binding of citrate and ADP, and the two effects together contribute to the NADH inhibition on the activity. These findings provide insights into the inhibitory mechanisms of the αγ heterodimer by NADH.
Organizational Affiliation:
School of Life Sciences, Shanghai University, 333 Nanchen Road, Shanghai, 200444, China.