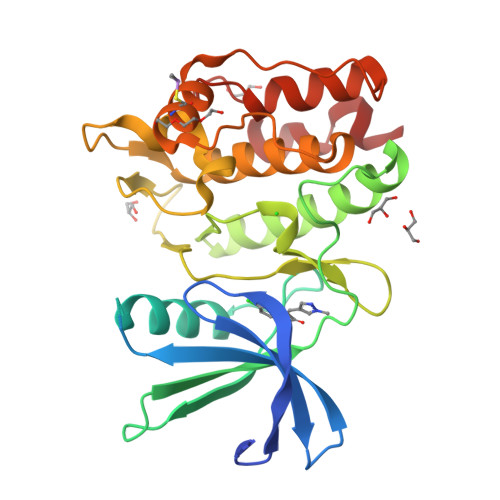
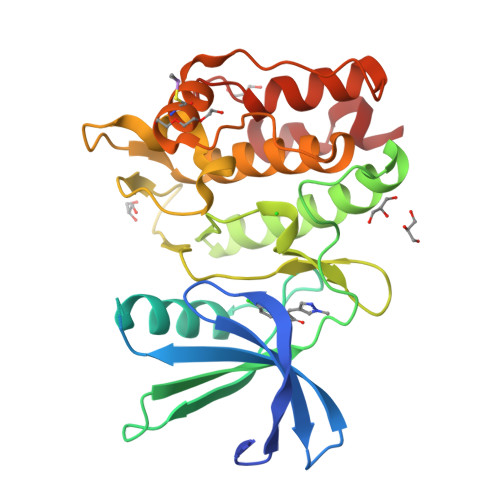
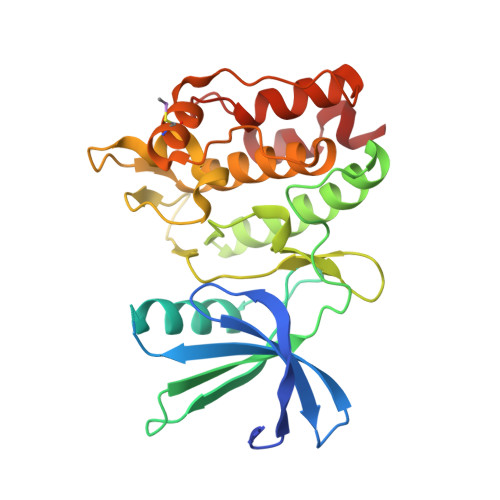
Protein ligand interaction analysis against new CaMKK2 inhibitors by use of X-ray crystallography and the fragment molecular orbital (FMO) method.
Takaya, D., Niwa, H., Mikuni, J., Nakamura, K., Handa, N., Tanaka, A., Yokoyama, S., Honma, T.(2020) J Mol Graph Model 99: 107599-107599
- PubMed: 32348940
- DOI: https://doi.org/10.1016/j.jmgm.2020.107599
- Primary Citation of Related Structures:
5YV8, 5YV9, 5YVA, 5YVB, 5YVC - PubMed Abstract:
CaMKK2 (calcium/calmodulin dependent protein kinase kinase 2) is a serine/threonine protein kinase that regulates phosphorylation of CaM kinases (CaMKs) such as CaMKI, CaMKIV, and AMP-activated protein kinase (AMPK). From a pathological perspective, CaMKK2 plays a role in obesity, diabetes, and prostate cancer. Therefore, CaMKK2 is an attractive target protein for drug design. Here, we tried to find new CaMKK2 inhibitors by using ligand-based and structure-based drug design approaches. From the in silico hit compounds, we identified new inhibitors by using a CaMKK2 kinase assay. We solved X-ray crystallography structures of the CaMKK2-inhibitor complexes and performed Fragment Molecular Orbital (FMO) calculations to analyze the protein-ligand interactions, identify the key residues in inhibitor binding, and quantitatively measure their contribution. We experimentally determined five CaMKK2-inhibitor structures and calculated the binding energies of the inhibitors by the FMO method plus MM-PBSA (Molecular Mechanics Poisson-Boltzmann Surface Area) approach. The results showed a high correlation (R = -0.89) between experimentally measured inhibitory activity (pIC 50 ) and the predicted ligand binding energy. We then quantitatively evaluated the contribution of each binding site residue in CaMKK2 by the IFIE (Inter-fragment Interaction Energy)/PIEDA (Pair Interaction Energy Decomposition Analysis) method. The IFIE values indicated that Lys194 and Glu236, which formed hydrogen bonds with the carboxylate groups of the inhibitors, were key residues for ligand binding. PIEDA revealed that the dispersion interaction of inhibitors with hydrophobic residues, such as Ile171, Phe267, and Leu319, contributed highly to ligand binding; we considered that this was due to CH-π interactions with methoxy groups and/or aromatic rings contained in our CaMKK2 inhibitor. These results from the quantitative interaction analysis by the FMO method are useful not only for future CaMMK2 inhibitor development but for application of the FMO method to in silico drug design.
Organizational Affiliation:
RIKEN Center for Biosystems Dynamics Research, 1-7-22 Suehiro-cho, Tsurumi-ku, Yokohama, Kanagawa, 230-0045, Japan; RIKEN Center for Life Science Technologies, 1-7-22 Suehiro-cho, Tsurumi-ku, Yokohama, 230-0045, Japan; RIKEN Systems and Structural Biology Center, 1-7-22 Suehiro-cho, Tsurumi, Yokohama, 230-0045, Japan.