RanL182A in complex with RanBP1-CRM1
Sun, Q., Zhang, Y.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| GTP-binding nuclear protein Ran | 216 | Homo sapiens | Mutation(s): 1 Gene Names: RAN, ARA24, OK/SW-cl.81 EC: 3.6.5 | 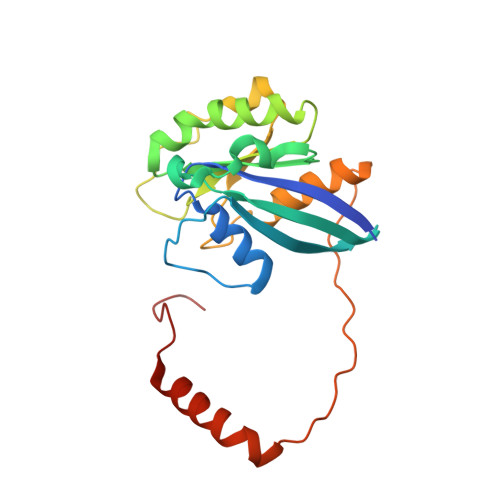 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P62826 (Homo sapiens) Explore P62826 Go to UniProtKB: P62826 | |||||
PHAROS: P62826 GTEx: ENSG00000132341 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P62826 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Ran-specific GTPase-activating protein 1 | 140 | Saccharomyces cerevisiae S288C | Mutation(s): 0 Gene Names: YRB1, CST20, HTN1, SFO1, YDR002W, YD8119.08 | 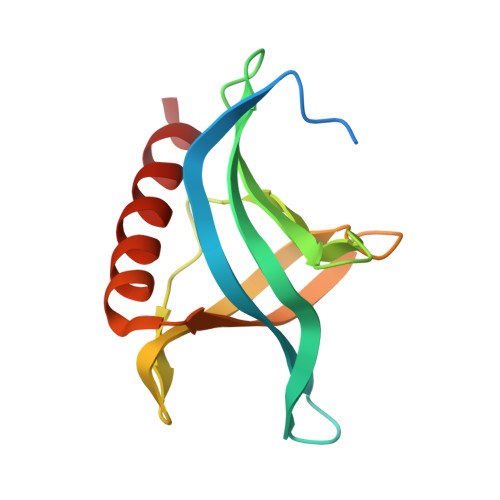 | |
UniProt | |||||
Find proteins for P41920 (Saccharomyces cerevisiae (strain ATCC 204508 / S288c)) Explore P41920 Go to UniProtKB: P41920 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P41920 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Exportin-1,Exportin-1 | 1,023 | Saccharomyces cerevisiae S288C | Mutation(s): 4 Gene Names: CRM1, KAP124, XPO1, YGR218W, G8514 | 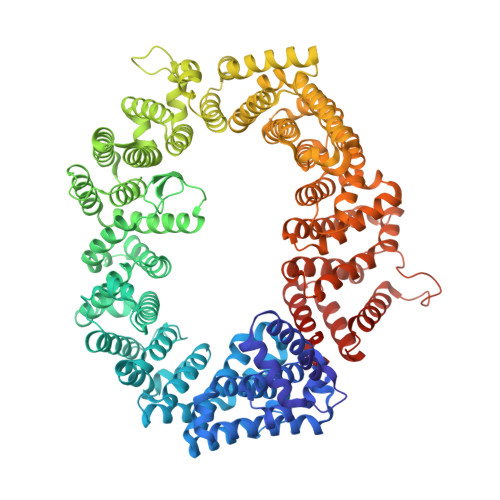 | |
UniProt | |||||
Find proteins for P30822 (Saccharomyces cerevisiae (strain ATCC 204508 / S288c)) Explore P30822 Go to UniProtKB: P30822 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P30822 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 5 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| GTP Query on GTP | D [auth A] | GUANOSINE-5'-TRIPHOSPHATE C10 H16 N5 O14 P3 XKMLYUALXHKNFT-UUOKFMHZSA-N |  | ||
| GOL Query on GOL | H [auth C] | GLYCEROL C3 H8 O3 PEDCQBHIVMGVHV-UHFFFAOYSA-N |  | ||
| EDO Query on EDO | F [auth A] | 1,2-ETHANEDIOL C2 H6 O2 LYCAIKOWRPUZTN-UHFFFAOYSA-N |  | ||
| CL Query on CL | I [auth C], J [auth C], K [auth C], L [auth C] | CHLORIDE ION Cl VEXZGXHMUGYJMC-UHFFFAOYSA-M |  | ||
| MG Query on MG | E [auth A], G [auth A] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 105.259 | α = 90 |
| b = 105.259 | β = 90 |
| c = 307.425 | γ = 90 |
| Software Name | Purpose |
|---|---|
| HKL-2000 | data scaling |
| PHENIX | refinement |
| PDB_EXTRACT | data extraction |
| HKL-2000 | data reduction |
| MOLREP | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| NSFC | China | 81502629 |