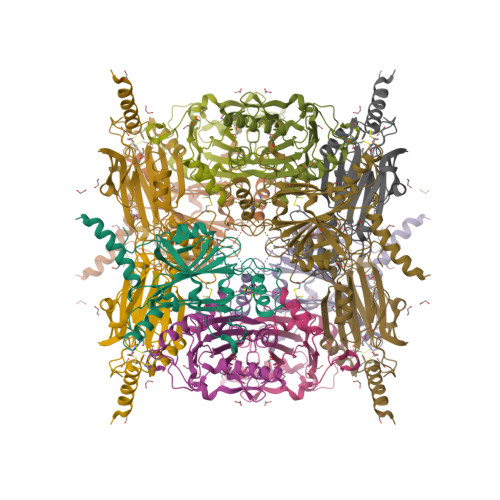
Supramolecular protein cages constructed from a crystalline protein matrix
Negishi, H., Abe, S., Yamashita, K., Hirata, K., Niwase, K., Boudes, M., Coulibaly, F., Mori, H., Ueno, T.(2018) Chem Commun (Camb) 54: 1988-1991
- PubMed: 29405208
- DOI: https://doi.org/10.1039/c7cc08689j
- Primary Citation of Related Structures:
5YR1, 5YR9, 5YRA, 5YRB, 5YRC, 5YRD - PubMed Abstract:
Protein crystals are formed via ordered arrangements of proteins, which assemble to form supramolecular structures. Here, we show a method for the assembly of supramolecular protein cages within a crystalline environment. The cages are stabilized by covalent cross-linking allowing their release via dissolution of the crystal. The high stability of the desiccated protein crystals allows cages to be constructed.
Organizational Affiliation:
School of Life Science and Technology, Tokyo Institute of Technology, Nagatsuta-cho, Midori-ku, Yokohama 226-8501, Japan. saabe@bio.titech.ac.jp tueno@bio.titech.ac.jp.