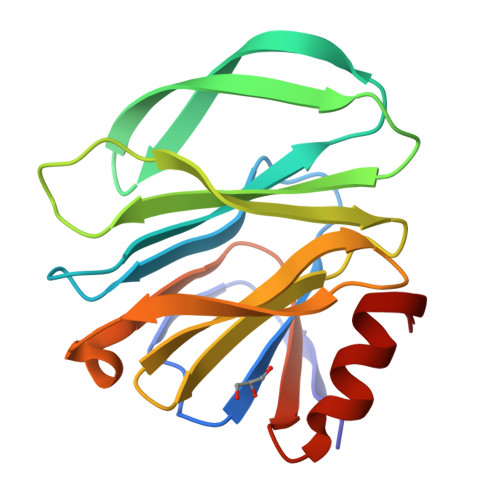
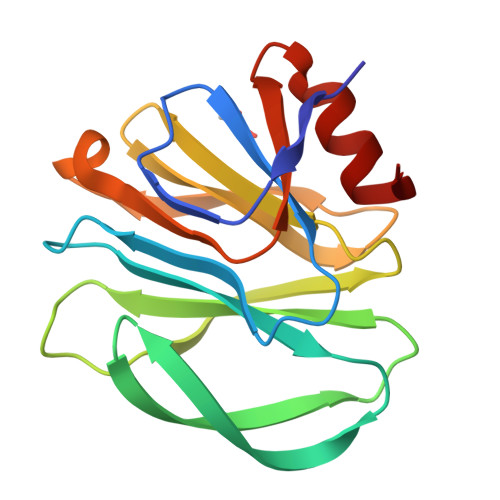
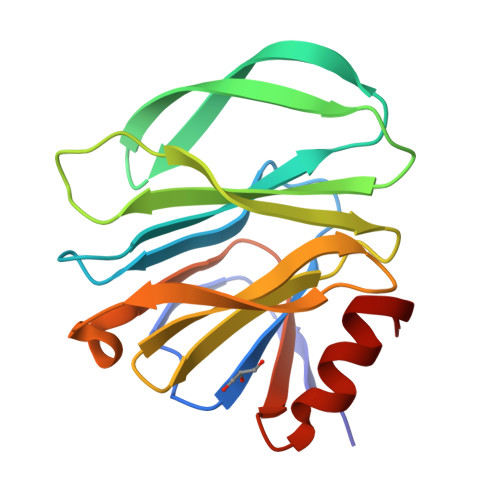
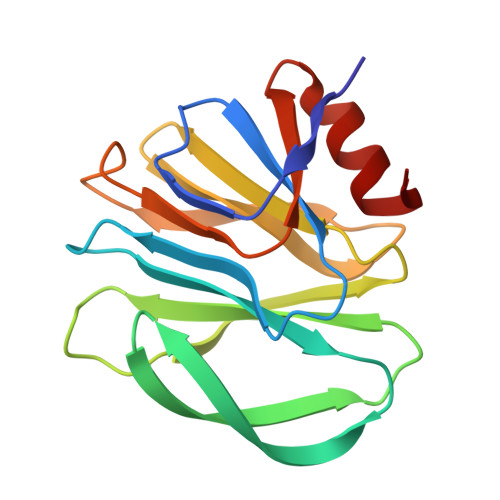
Glycan Binding Specificity and Mechanism of Human and Porcine P[6]/P[19] Rotavirus VP8*s.
Sun, X., Li, D., Qi, J., Chai, W., Wang, L., Wang, L., Peng, R., Wang, H., Zhang, Q., Pang, L., Kong, X., Wang, H., Jin, M., Gao, G.F., Duan, Z.(2018) J Virol 92
- PubMed: 29720519
- DOI: https://doi.org/10.1128/JVI.00538-18
- Primary Citation of Related Structures:
5YMS, 5YMT, 5YMU - PubMed Abstract:
Rotaviruses (RVs), which cause severe gastroenteritis in infants and children, recognize glycan ligands in a genotype-dependent manner via the distal VP8* head of the spike protein VP4. However, the glycan binding mechanisms remain elusive for the P[II] genogroup RVs, including the widely prevalent human RVs (P[8], P[4], and P[6]) and a rare P[19] RV. In this study, we characterized the glycan binding specificities of human and porcine P[6]/P[19] RV VP8*s and found that the P[II] genogroup RV VP8*s could commonly interact with mucin core 2, which may play an important role in RV evolution and cross-species transmission. We determined the first P[6] VP8* structure, as well as the complex structures of human P[19] VP8*, with core 2 and lacto- N -tetraose (LNT). A glycan binding site was identified in human P[19] VP8*. Structural superimposition and sequence alignment revealed the conservation of the glycan binding site in the P[II] genogroup RV VP8*s. Our data provide significant insight into the glycan binding specificity and glycan binding mechanism of the P[II] genogroup RV VP8*s, which could help in understanding RV evolution, transmission, and epidemiology and in vaccine development. IMPORTANCE Rotaviruses (RVs), belonging to the family Reoviridae , are double-stranded RNA viruses that cause acute gastroenteritis in children and animals worldwide. Depending on the phylogeny of the VP8* sequences, P[6] and P[19] RVs are grouped into genogroup II, together with P[4] and P[8], which are widely prevalent in humans. In this study, we characterized the glycan binding specificities of human and porcine P[6]/P[19] RV VP8*s, determined the crystal structure of P[6] VP8*, and uncovered the glycan binding pattern in P[19] VP8*, revealing a conserved glycan binding site in the VP8*s of P[II] genogroup RVs by structural superimposition and sequence alignment. Our data suggested that mucin core 2 may play an important role in P[II] RV evolution and cross-species transmission. These data provide insight into the cell attachment, infection, epidemiology, and evolution of P[II] genogroup RVs, which could help in developing control and prevention strategies against RVs.
Organizational Affiliation:
Key Laboratory of Medical Virology and Viral Diseases, Ministry of Health of the People's Republic of China, Beijing, China.