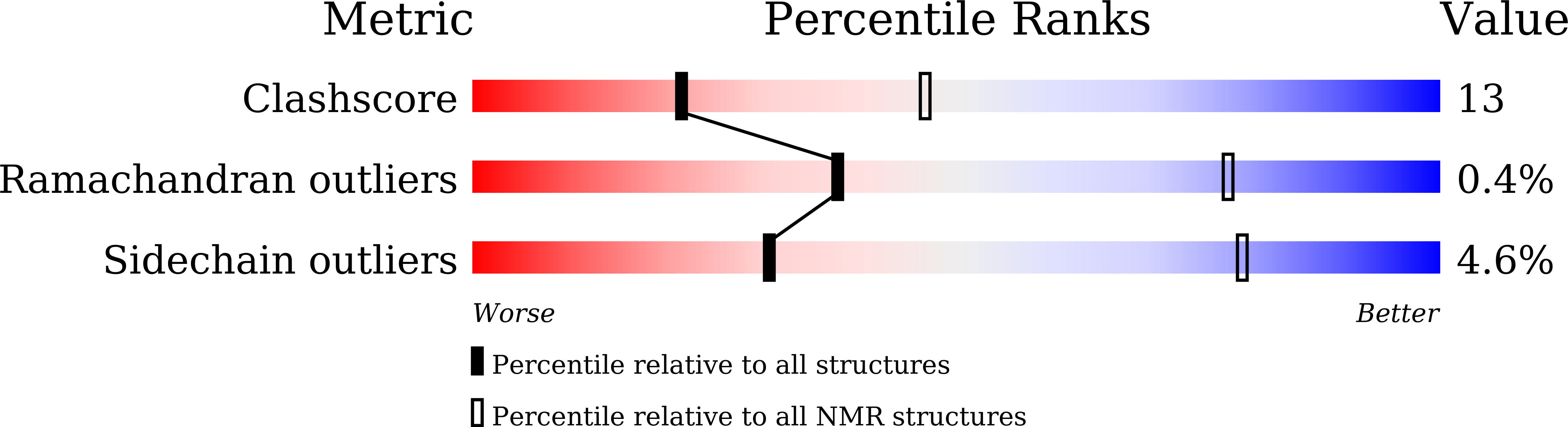
Integrative Model to Coordinate the Oligomerization and Aggregation Mechanisms of CCL5.
Chen, Y.C., Chen, S.P., Li, J.Y., Chen, P.C., Lee, Y.Z., Li, K.M., Zarivach, R., Sun, Y.J., Sue, S.C.(2020) J Mol Biol 432: 1143-1157
- PubMed: 31931012
- DOI: https://doi.org/10.1016/j.jmb.2019.12.049
- Primary Citation of Related Structures:
5YAM, 6AEZ - PubMed Abstract:
CC-type chemokine ligand 5 (CCL5) is involved in the pathogenesis of many inflammatory conditions. Under physiological conditions, CCL5 oligomerization and aggregation are considered to be responsible for its inflammatory properties. The structural basis of CCL5 oligomerization remains controversial because the current oligomer models contain no consensus interactions. In this study, NMR and biophysical analyses proposed evidence that the CC-type CCL5 dimer acts as the basic unit to constitute the oligomer and that CCL5 oligomerizes alternatively through E66-K25 and E66-R44/K45 interactions. In addition, a newly determined trimer structure, constituted by CCL5 and the E66S mutant, reported an interfacial interaction through the N-terminal 12 FAY 14 sequence. The interaction contributes to CCL5 aggregation and precipitation but not to oligomerization. In accordance with the observations, an integrative model explains the CCL5 oligomerization and aggregation mechanism in which CCL5 assembly consists of two types of dimer-dimer interactions and one aggregation mechanism. For full-length CCL5, the molecular accumulation triggers oligomerization through the E66-K25 and E66-R44/K45 interactions, and the 12 FAY 14 interaction acts as a secondary effect to derive aggregation and precipitation. In contrast, the E66-R44/K45 interaction might dominate in CCL5 N-terminal truncations, and the interaction would lead to the filament-like formation in solution.
Organizational Affiliation:
Institute of Bioinformatics and Structural Biology, National Tsing Hua University, Hsinchu, 30013, Taiwan.