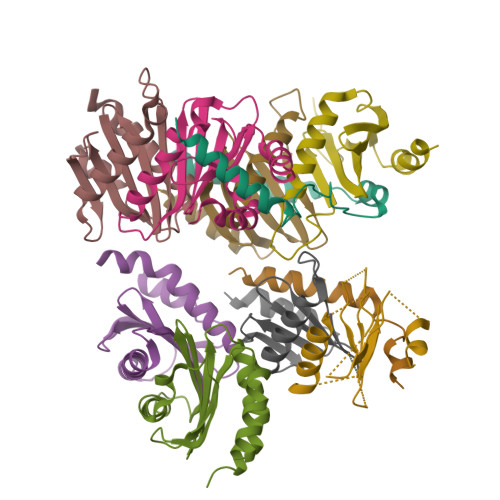
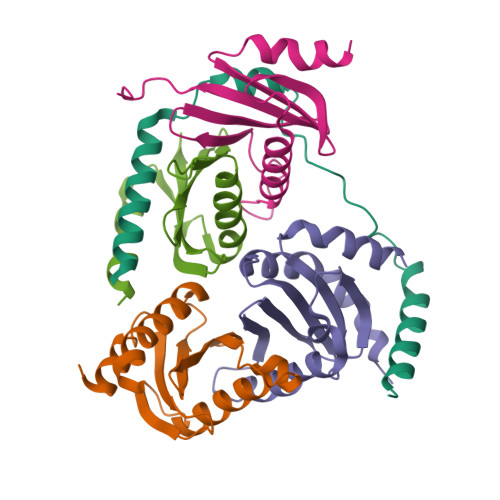
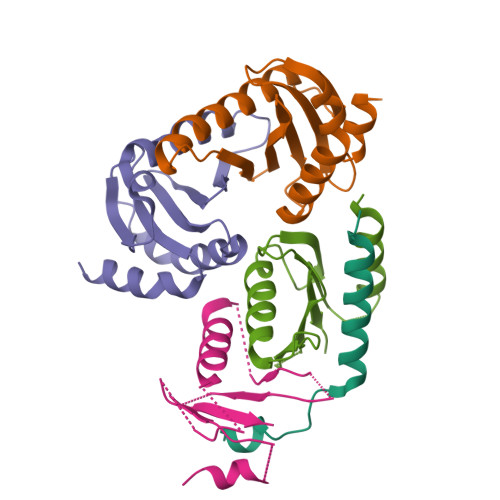
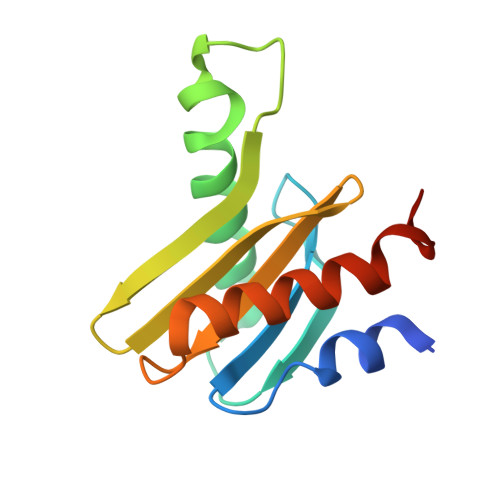
Structural basis for Ragulator functioning as a scaffold in membrane-anchoring of Rag GTPases and mTORC1
Zhang, T., Wang, R., Wang, Z., Wang, X., Wang, F., Ding, J.(2017) Nat Commun 8: 1394-1394
- PubMed: 29123114
- DOI: https://doi.org/10.1038/s41467-017-01567-4
- Primary Citation of Related Structures:
5Y38, 5Y39, 5Y3A - PubMed Abstract:
Amino acid-dependent activation of the mechanistic target of rapamycin complex 1 (mTORC1) is mediated by Rag GTPases, which are recruited to the lysosome by the Ragulator complex consisting of p18, MP1, p14, HBXIP and C7orf59; however, the molecular mechanism is elusive. Here, we report the crystal structure of Ragulator, in which p18 wraps around the MP1-p14 and C7orf59-HBXIP heterodimers and the interactions of p18 with MP1, C7orf59, and HBXIP are essential for the assembly of Ragulator. There are two binding sites for the Roadblock domains of Rag GTPases: helix α1 of p18 and the two helices side of MP1-p14. The interaction of Ragulator with Rag GTPases is required for their cellular co-localization and can be competitively inhibited by C17orf59. Collectively, our data indicate that Ragulator functions as a scaffold to recruit Rag GTPases to lysosomal membrane in mTORC1 signaling.
Organizational Affiliation:
State Key Laboratory of Molecular Biology, National Center for Protein Science Shanghai, Shanghai Science Research Center, CAS Center for Excellence in Molecular Cell Science, Institute of Biochemistry and Cell Biology, Shanghai Institutes for Biological Sciences, University of Chinese Academy of Sciences, Chinese Academy of Sciences, 320 Yue-Yang Road, Shanghai, 200031, China.