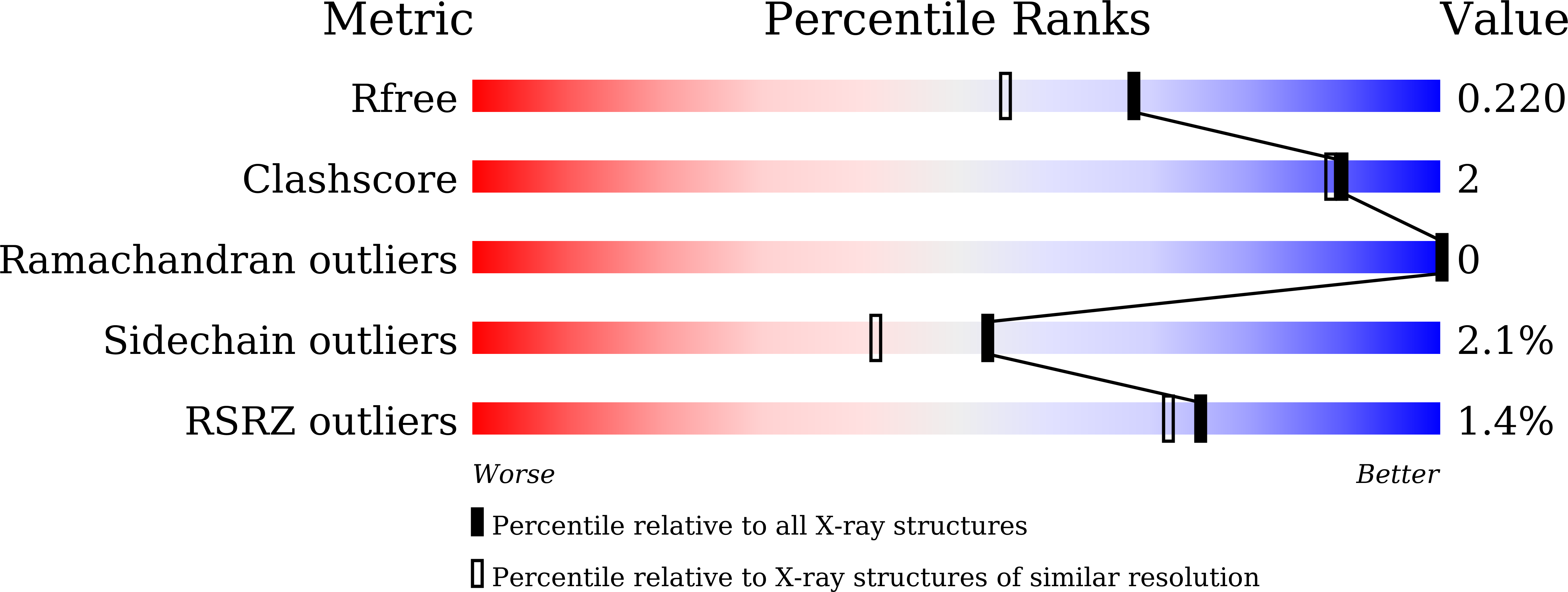
The crystal structure and catalytic mechanism of hydroxynitrile lyase from passion fruit, Passiflora edulis
Motojima, F., Nuylert, A., Asano, Y.(2018) FEBS J 285: 313-324
- PubMed: 29155493
- DOI: https://doi.org/10.1111/febs.14339
- Primary Citation of Related Structures:
5XZQ, 5XZT, 5Y02 - PubMed Abstract:
Hydroxynitrile lyases (HNLs) are enzymes used in the synthesis of chiral cyanohydrins. The HNL from Passiflora edulis (PeHNL) is R-selective and is the smallest HNL known to date. The crystal structures of PeHNL and its C-terminal peptide depleted derivative were determined by molecular replacement method using the template structure of a heat stable protein, SP1, from Populus tremula at 2.8 and 1.8 Å resolution, respectively. PeHNL belongs to dimeric α+β barrel superfamily consisting of a central β-barrel in the middle of a dimer. The structure of PeHNL complexed with (R)-mandelonitrile ((R)-MAN) was also determined. The hydroxyl group of (R)-MAN forms hydrogen bonds with His8 and Tyr30 in the active site, whereas the nitrile group is oriented toward the carboxyl group of Glu54, unlike other HNLs, where it interacts with basic residues typically. The results of mutational analysis indicate that the catalytic dyad of His8-Asn101 is critical for the enzymatic reaction. The length of the hydrogen bond between His-Nδ1 and Asn101-Oδ1 is short in the PeHNL-(R)-MAN complex (~ 2.6 Å), which would increase the basicity of His8 to abstract a proton from the hydroxyl group of (R)-MAN. The cyanide ion released from the nitrile group abstracts a proton from the protonated His8 to generate a hydrogen cyanide. Thus, the His8 in the active site of PeHNL acts both as a general acid and a general base in the reaction. EC 4.1.2.10 DATABASE: Structural data are available in PDB database under the accession numbers 5XZQ, 5XZT, and 5Y02.
Organizational Affiliation:
Biotechnology Research Center and Department of Biotechnology, Toyama Prefectural University, Imizu, Japan.