25 S-Adamantyl-23-yne-26,27-dinor-1 alpha ,25-dihydroxyvitamin D3: Synthesis, Tissue Selective Biological Activities, and X-ray Crystal Structural Analysis of Its Vitamin D Receptor Complex.
Otero, R., Ishizawa, M., Numoto, N., Ikura, T., Ito, N., Tokiwa, H., Mourino, A., Makishima, M., Yamada, S.(2018) J Med Chem 61: 6658-6673
- PubMed: 29989817
- DOI: https://doi.org/10.1021/acs.jmedchem.8b00427
- Primary Citation of Related Structures:
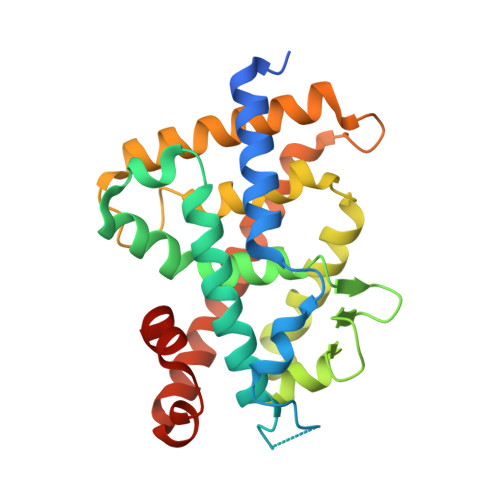
5XZF, 5XZH - PubMed Abstract:
Both 25 R- and 25 S-25-adamantyl-23-yne-26,27-dinor-1α,25-dihydroxyvitamin D 3 (4a and 4b) were stereoselectively synthesized by a Pd(0)-catalyzed ring closure and Suzuki-Miyaura coupling between enol-triflate 7 and alkenyl-boronic ester 8. The 25 S isomer (4b) showed high vitamin D receptor (VDR) affinity (50% of that of the natural hormone 1α,25-dihydroxyvitamin D 3 , 1) and transactivation potency (kidney HEK293, 90%). In endogenous gene expression, it showed high cell-type selectivity for kidney cells (HEK293, CYP24A1 160% of 1), bone cells (MG63, osteocalcin 64%), and monocytes (U937, CAMP 96%) over intestine (SW480, CYP24A1 8%) and skin (HaCaT, CYP24A1 7%) cells. The X-ray crystal structural analysis of 4b in complex with rat VDR-ligand binding domain (LBD) showed the highest Cα positional shift from the 1/VDR-LBD complex at helix 11. Helix 11 of the 4b and 1 VDR-LBD complexes also showed significant differences in surface properties. These results suggest that 4b should be examined further as another candidate for a mild preventive osteoporosis agent.
Organizational Affiliation:
Departamento de Química Orgánica, Laboratorio de Investigación Ignacio Ribas , Universidad de Santiago de Compostela , 15782 Santiago de Compostela , Spain.