Hydrophobic patches on SMAD2 and SMAD3 determine selective binding to cofactors
Miyazono, K.I., Moriwaki, S., Ito, T., Kurisaki, A., Asashima, M., Tanokura, M.(2018) Sci Signal 11
- PubMed: 29588413
- DOI: https://doi.org/10.1126/scisignal.aao7227
- Primary Citation of Related Structures:
5XOC, 5XOD - PubMed Abstract:
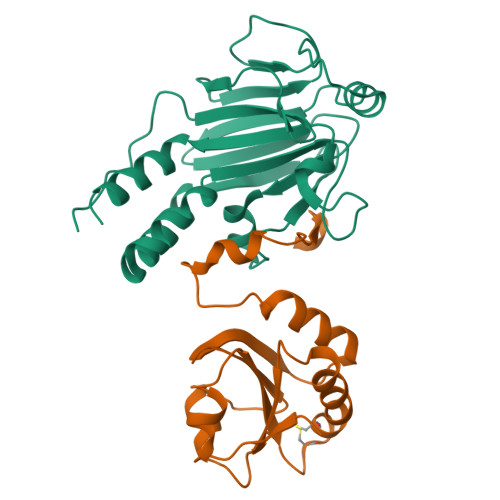
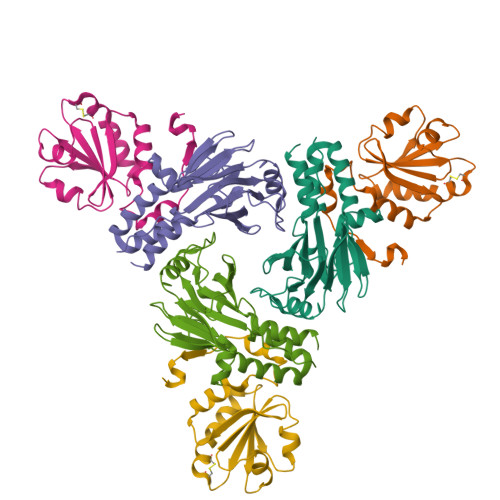
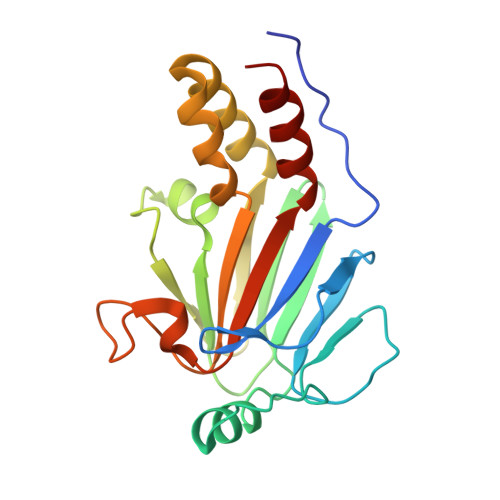
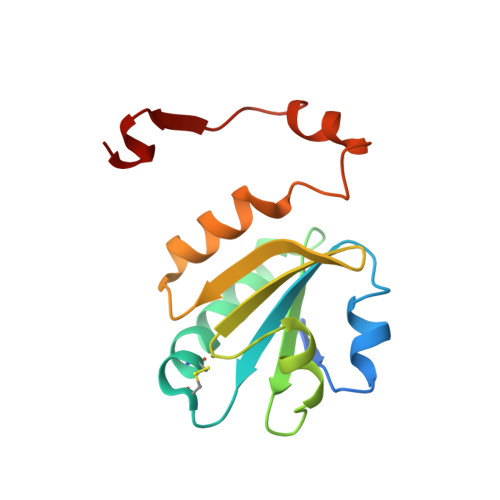
The transforming growth factor-β (TGF-β) superfamily of cytokines regulates various biological processes, including cell proliferation, immune responses, autophagy, and senescence. Dysregulation of TGF-β signaling causes various diseases, such as cancer and fibrosis. SMAD2 and SMAD3 are core transcription factors involved in TGF-β signaling, and they form heterotrimeric complexes with SMAD4 (SMAD2-SMAD2-SMAD4, SMAD3-SMAD3-SMAD4, and SMAD2-SMAD3-SMAD4) in response to TGF-β signaling. These heterotrimeric complexes interact with cofactors to control the expression of TGF-β-dependent genes. SMAD2 and SMAD3 may promote or repress target genes depending on whether they form complexes with other transcription factors, coactivators, or corepressors; therefore, the selection of specific cofactors is critical for the appropriate activity of these transcription factors. To reveal the structural basis by which SMAD2 and SMAD3 select cofactors, we determined the crystal structures of SMAD3 in complex with the transcription factor FOXH1 and SMAD2 in complex with the transcriptional corepressor SKI. The structures of the complexes show that the MAD homology 2 (MH2) domains of SMAD2 and SMAD3 have multiple hydrophobic patches on their surfaces. The cofactors tether to various subsets of these patches to interact with SMAD2 and SMAD3 in a cooperative or competitive manner to control the output of TGF-β signaling.
Organizational Affiliation:
Laboratory of Basic Science on Healthy Longevity, Department of Applied Biological Chemistry, Graduate School of Agricultural and Life Sciences, The University of Tokyo, Tokyo 113-8657, Japan.