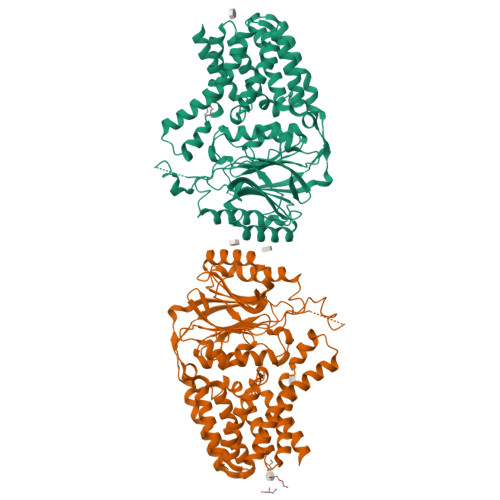
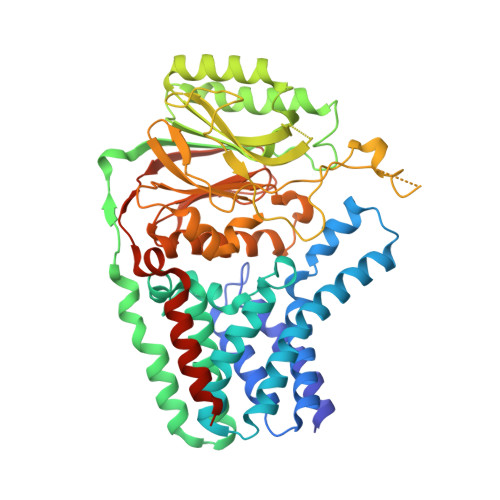
Crystal structure of E. coli apolipoprotein N-acyl transferase
Lu, G., Xu, Y., Zhang, K., Xiong, Y., Li, H., Cui, L., Wang, X., Lou, J., Zhai, Y., Sun, F., Zhang, X.C.(2017) Nat Commun 8: 15948-15948
- PubMed: 28885614
- DOI: https://doi.org/10.1038/ncomms15948
- Primary Citation of Related Structures:
5XHQ - PubMed Abstract:
In Gram-negative bacteria, lipid modification of proteins is catalysed in a three-step pathway. Apolipoprotein N-acyl transferase (Lnt) catalyses the third step in this pathway, whereby it transfers an acyl chain from a phospholipid to the amine group of the N-terminal cysteine residue of the apolipoprotein. Here, we report the 2.6-Å crystal structure of Escherichia coli Lnt. This enzyme contains an exo-membrane nitrilase domain fused to a transmembrane (TM) domain. The TM domain of Lnt contains eight TM helices which form a membrane-embedded cavity with a lateral opening and a periplasmic exit. The nitrilase domain is located on the periplasmic side of the membrane, with its catalytic cavity connected to the periplasmic exit of the TM domain. An amphipathic lid loop from the nitrilase domain interacts with the periplasmic lipid leaflet, forming an interfacial entrance from the lipid bilayer to the catalytic centre for both the lipid donor and acceptor substrates.
Organizational Affiliation:
National Laboratory of Macromolecules, Institute of Biophysics, CAS Center for Excellence in Biomacromolecules, Chinese Academy of Sciences, 15 Datun Road, Beijing 100101, China.